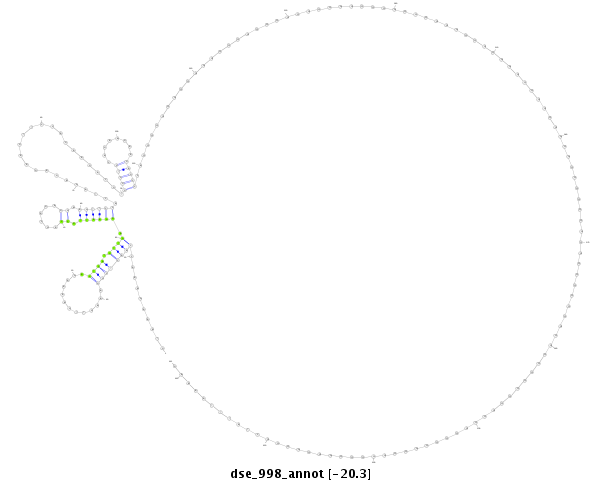
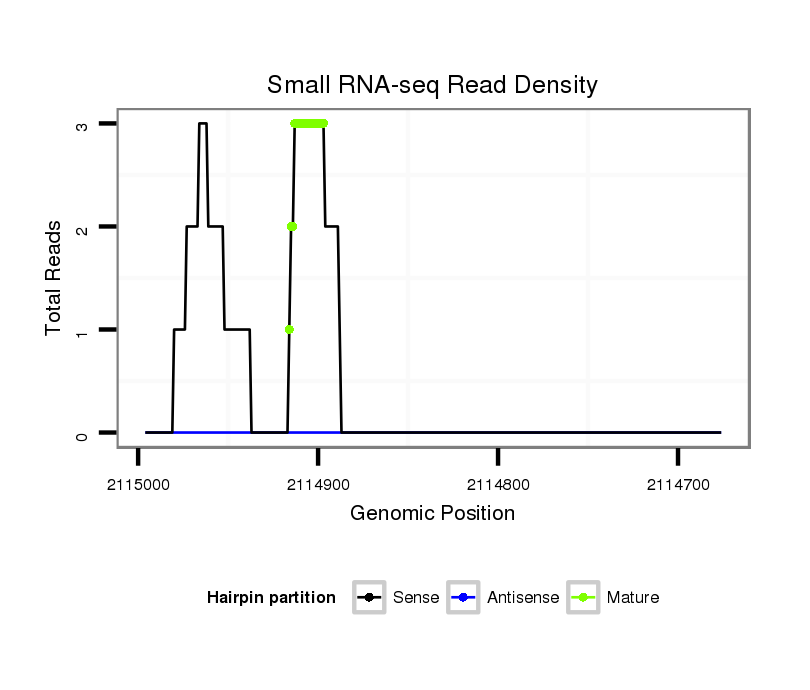
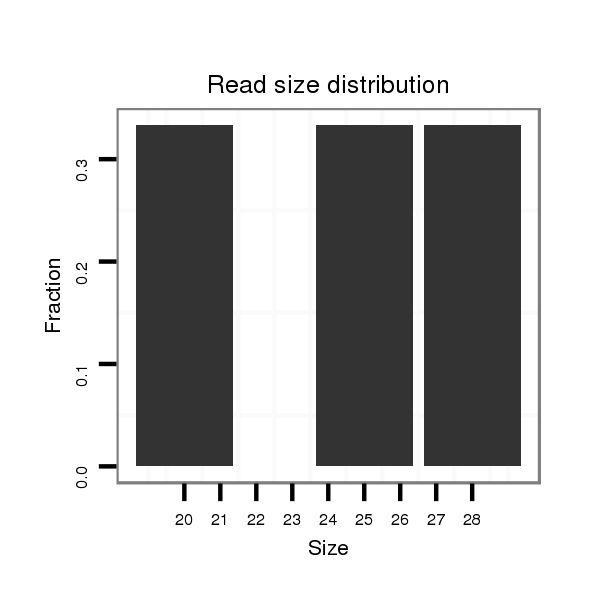
ID:dse_998 |
Coordinate:scaffold_2:2114726-2114946 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -20.3 | -20.2 | -20.2 |
 |
 |
 |
CDS [Dsec\GM14145-cds]; exon [dsec_GLEANR_14941:3]; CDS [Dsec\GM14145-cds]; exon [dsec_GLEANR_14941:2]; intron [Dsec\GM14145-in]
No Repeatable elements found
| ##################################################-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CTGCTGAGGACCAGCAACTAGTCGTTGAAGAGAGCAAAAAGGTTAAAAAAGTAAAGAAACCAACTCGGAAAGTTGAAAAGACTGATGTTGAGGAATTGCCAGCAGAGGAAGTTCCACTAGAGAGAAGTTCCAGAAGACGTGGCTCCAGAAGAAGAGCCCANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCTATTTAAAAGACCGAAATAGAAAGAGAAGAAGACCAACCAGAAGAGGAAGTTCCACAAGA **************************************************..........((((((((.............)))).)))).((((((.((......)).)))))).........................(((((........))))).................................................................................................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V113 male body |
M054 female body |
V115 head |
V114 embryo |
|---|---|---|---|---|---|---|---|---|---|
| .............................................................................AAGACCAATGATGAGGAATTG............................................................................................................................................................................................................................... | 21 | 3 | 1 | 24.00 | 24 | 20 | 4 | 0 | 0 |
| ...........................................................................AAAAGACCAATGATGAGGAAT................................................................................................................................................................................................................................. | 21 | 3 | 4 | 9.75 | 39 | 34 | 1 | 4 | 0 |
| ...........................................................................AAAAGACCAATGATGAGGAATT................................................................................................................................................................................................................................ | 22 | 3 | 1 | 6.00 | 6 | 6 | 0 | 0 | 0 |
| ............................................................................AAAGACCAATGATGAGGAATT................................................................................................................................................................................................................................ | 21 | 3 | 3 | 4.33 | 13 | 9 | 2 | 2 | 0 |
| ...........................................................................AAAAGACCAATGATGAGGAATTG............................................................................................................................................................................................................................... | 23 | 3 | 1 | 3.00 | 3 | 1 | 1 | 1 | 0 |
| ..........................................................................GAAAAGACCAATGATGAGGAAT................................................................................................................................................................................................................................. | 22 | 3 | 3 | 2.33 | 7 | 6 | 1 | 0 | 0 |
| ..........................................................................GAAAAGACCAATGATGAGGAATTG............................................................................................................................................................................................................................... | 24 | 3 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ..........................................................................GAAAAGACCAATGATGAGGAA.................................................................................................................................................................................................................................. | 21 | 3 | 7 | 1.43 | 10 | 7 | 3 | 0 | 0 |
| ................ACTAGTCGTTGAAGAGAGC.............................................................................................................................................................................................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ..........................................................................GAAAAGACCAATGATGAGGAATT................................................................................................................................................................................................................................ | 23 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................ACTGATGTTGAGGAATTGCC............................................................................................................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................AAAGACTAATGATGAGGAATT................................................................................................................................................................................................................................ | 21 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .................................................................................CTGATGTTGAGGAATTGCCAGCAGAGGA.................................................................................................................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................GTTGAAGAGAGCAAAAAGGTT..................................................................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ...................................................................................GATGTTGAGGAATTGCCAGCAGAGG..................................................................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................AAAGACCAATGATGAGGAATTG............................................................................................................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..............................AGAGCAAAAAGGTTAAAAAAGTAAAGAAA...................................................................................................................................................................................................................................................................... | 29 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................AAGACTAATGATGAGGAATTG............................................................................................................................................................................................................................... | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................AAAGACCAATGATGAGGAAT................................................................................................................................................................................................................................. | 20 | 3 | 13 | 0.69 | 9 | 8 | 1 | 0 | 0 |
| .............................................................................AAGACCAATGATGAGGAATT................................................................................................................................................................................................................................ | 20 | 3 | 9 | 0.56 | 5 | 2 | 3 | 0 | 0 |
| ...........................................................................AAAAGACCAATGATGAGGAA.................................................................................................................................................................................................................................. | 20 | 3 | 20 | 0.35 | 7 | 7 | 0 | 0 | 0 |
| ..................................................................................TAATGATGAGGAATTGGCAGC.......................................................................................................................................................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................AAAGACAAATGATGAGGAATT................................................................................................................................................................................................................................ | 21 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................AAGCAGAAAGAGAAGAGGACC......................... | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 |
| ......................................AAGGTGAAAAAAGTAAAAAAA...................................................................................................................................................................................................................................................................... | 21 | 2 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................CGTGGTTCCTGAAGAAGAC..................................................................................................................................................................... | 19 | 3 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................................AGACCAATGATGAGGAATTGC.............................................................................................................................................................................................................................. | 21 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................AAATACCGACATAGAAAGC................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .................................................GGAAAAGAAACCAACTGGGA............................................................................................................................................................................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ...............................................................................GACCAATGATGAGGAATTG............................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................CAAATAGAGAGGGAAGAAGA........................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
|
GACGACTCCTGGTCGTTGATCAGCAACTTCTCTCGTTTTTCCAATTTTTTCATTTCTTTGGTTGAGCCTTTCAACTTTTCTGACTACAACTCCTTAACGGTCGTCTCCTTCAAGGTGATCTCTCTTCAAGGTCTTCTGCACCGAGGTCTTCTTCTCGGGTNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNGATAAATTTTCTGGCTTTATCTTTCTCTTCTTCTGGTTGGTCTTCTCCTTCAAGGTGTTCT
**************************************************..........((((((((.............)))).)))).((((((.((......)).)))))).........................(((((........))))).................................................................................................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M054 female body |
V113 male body |
|---|---|---|---|---|---|---|---|
| .............................................................................TTCTGGTTACTACTCCTTAAC............................................................................................................................................................................................................................... | 21 | 3 | 1 | 4.00 | 4 | 4 | 0 |
| ............................................................................TTTCTGGTTACTACTCCTTAAC............................................................................................................................................................................................................................... | 22 | 3 | 1 | 3.00 | 3 | 1 | 2 |
| ............................................................................TTTCTGGTTACTACTCCTTAA................................................................................................................................................................................................................................ | 21 | 3 | 3 | 2.33 | 7 | 7 | 0 |
| ...........................................................................TTTTCTGGTTACTACTCCTTA................................................................................................................................................................................................................................. | 21 | 3 | 4 | 1.75 | 7 | 5 | 2 |
| .............................................................................TTCTGGTTACTACTCCTTAA................................................................................................................................................................................................................................ | 20 | 3 | 9 | 0.33 | 3 | 2 | 1 |
| ..........................................................................CTTTTCTGGTTACTACTCCTT.................................................................................................................................................................................................................................. | 21 | 3 | 7 | 0.29 | 2 | 2 | 0 |
| ...........................................ATTTATTCATTTCTTTGAT................................................................................................................................................................................................................................................................... | 19 | 2 | 20 | 0.05 | 1 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSec2 | scaffold_2:2114676-2114996 - | dse_998 | C---TGCTGAGGACCAGCAACTAGTCGT---TGAAGAGAGCAAAAAGGTTAAAAAAGTAAAGAAACCAACTCGGAAAGTT---GAAA---------AGACT-------GATGTTGAGGAAT---------TGCCAGCAGAGGAAGTTCCACTAGAGA--------------GAAGTTCCAGAAGACGTGGCTCCAGAAGAAGAGCCCANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCTATTTAAAAGACCGAAATAGAAAGAGAAG---------AAGACCAACCAGAAGAGGAAGTTCC-ACAA---G--------A |
| droSim2 | 3l:1987132-1987305 - | C---TGCTGAGGACCAGCAACTAGTCGT---TGAAGAGAGCAAAAAGGTTAAAAAAGTAAAGAAACCAACTCGGAAAGTT---GAAA---------AGACT-------GATGTTGAGGAAT---------TGCCAGCAGAGGAAGTTCCAGCAGAGGAAGTTATAGTTGAGGAAGTTCCAGAAGAGGTGGCTCCAGAAGAA----------------------------------------------------------------------------------------------------------------------------------GAGC----------------------------------------------------CCA | |
| dm3 | chr3L:2084023-2084196 - | C---TGCTGAGGACCAGCAACTAGTCGT---TGAAGAGAGCAAAAAGGTTAAAAAAGTAAAGAAACCAACTGGGACAGTT---GAAA---------AGACT-------GATGTTGAGGAAT---------TGCCAGGAGAGGAAGTTCCAGTAGAGGAAGTTCCAGTAGAGGAAGTTCCAGAAGACGTGGCTCCAGAAGAA----------------------------------------------------------------------------------------------------------------------------------GAGC----------------------------------------------------TCA | |
| droEre2 | scaffold_4784:2082905-2083059 - | C---TGTTGAGAAGCAGCAATCAGTTGT---TGAAGAAAGCGAAAAGGTTAAAAAAGTAAAGAAACCAACACGAACGGTT---GAAA---------AGCTT-------GATGTCCAGGAAC---------TGCCAGAAGAGGATGTTCCGGAAAAGGAAA---------------TTCCAGT---------------GGAT----------------------------------------------------------------------------------------------------------------------------------GAGCCCA---------TCGAGGAGGAG-------------------------------- | |
| droYak3 | 3L:2042292-2042446 - | C---TGCTGACG---------TTGTTGT---CGAAGAGAGCAAACAGGTTAAAAAGGTAAAGAAACCAACTCGAACAGTT---GAAA---------AGACT-------GATGTTGAGGATC---------TGCCAGTAGAGGAAGTTCCAAA---------------------------------------------AGAT----------------------------------------------------------------------------------------------------------------------------------GAGCCCG---------TCGAGGAGCAGGAGGAGATCGTTGC-CCAA---GAAGA----A | |
| droEug1 | scf7180000409091:184379-184517 + | CGATCGTGGAAGATCAAGAAGAGGTTGT---TGAAGAGAAGAAAAAGGTCAAAAAGGCAAAGAAGCCCAAAGCAACCGTT---GAAA---------AGACT-------GATGTTGAAGAAC---------AGCCTGAAGAGGAAATTCCAGA---------------------------------------------AGAA----------------------------------------------------------------------------------------------------------------------------------GAGCTTA---------TTGAAGA------------------------------------ | |
| droBia1 | scf7180000302428:8811543-8811709 - | T---TGTTCAGGACCAGGAAGAGGTCGCTGTTGAAGAAAAACGCCGGGTCAGGAAGGTCAAGAAGCCCAAAACAGTGGTT---GAGA---------AGACAG----AAGATGTGGAGGAAC---------AGCCCGAAGATGATATCCCGCA---------------------------------------------GGAA----------------------------------------------------------------------------------------------------------------------------------GAAATCG---------CCGAAACGCAAGAATTGGTCGTGGA-AGAA---AA-------G | |
| droTak1 | scf7180000414400:15217-15382 - | CGATCGTGGAAGACCAGGAAGAGGTTGT---TGAAGAGAAGAAGAAGGTCAAGAGGGTAAGGAAGCCCAAACCAACCGTT---GAAA---------AGACT-------GATGTTGAAGAAC---------AGCCTGAGGAAGATATTCCGGA---------------------------------------------GGAG----------------------------------------------------------------------------------------------------------------------------------GAGGCCA---------TTGAAGAGCAAGAAGAGGTTGTAGA-ACAA---GAAG-----A | |
| droEle1 | scf7180000491249:3407322-3407484 + | CGGTCGTTCAGGATCAACAAGAGGTTGT---TGAGGAAAAGAAAAAGGTTAAAAAAGTAAAGAAGCCCAAGCAAACCGTT---GAAA---------ATACT-------GCTGTTGAGGAAC---------AGCCTGTAGAGGAAATTCCAGA---------------------------------------------AG-------------------------------------------------------------------------------------------------------------------------------------AGGTCA---------TTGAAGAACCTGAAGAGGTCATCGA-AGAA---CAAG-----A | |
| droRho1 | scf7180000779211:14920-15077 - | T---CGTAGAGGA------AGAGGTTGT---TGAAGTGAAGAAGAAGGTCAAAAAAGTAAAGAAACCAAAGCAACCCGTT---GAAA---------AGACT-------GATGTTGAAGAAC---------AGCTTGTAGAGGAAATTCCAGA---------------------------------------------AGAA----------------------------------------------------------------------------------------------------------------------------------GAGGTCA---------TCGACGAGCTAGAAGAGATCGTAGA-ACAA---GAAGA----A | |
| droFic1 | scf7180000453052:971953-972091 + | CACTCGACGAAGTGCAAGAAGAAGTTTT---GGAGGAGACGAAAAAGGTCAAAAAGGTTAAAAAACCCAAATTAACCGTT---GAAA---------AGACT-------GATGTTCAAGAAC---------AGCCACAAGAGGATATTCCAGA---------------------------------------------AGAA----------------------------------------------------------------------------------------------------------------------------------GAGATCA---------TCGAAGA------------------------------------ | |
| droKik1 | scf7180000302441:1719388-1719567 + | CGATTGTTGAAGAGCAACAAGAAGTTAT---TGAGGAGAAGAAAAAGGTCAAGAAGGTTAAAAAGCCCAAACCAACCGTT---GAAA---------AGACT-------GATGTCGAGGAACT---GGAACAGCCTGAAGAGGAGGTCCCAGAAGAGCAG---------------------------------------GTA----------------------------------------------------------------------------------------------------------------------------------GAGATTA---------CTGAAGAACAAGAAGAAGTCGTTGA-AGAA---AAGC---GCA | |
| droAna3 | scaffold_13337:741697-741843 + | C---CACGGAGGATCA------AATTGT---GGACGTAACCAAGAAGCGTGTTAAGAAAAAGAAACCAAAG------------GCAA---------AGACT-------CCTACAGAAGAATCGTTTGAGGAGCCTGTGGAAGATGT------------------------------------------------------G----------------------------------------------------------------------------------------------------------------------------------GAAGAAT------TTGAAGACA----CGCAGCCTCAGCCAGATGAA------GAG---C | |
| droBip1 | scf7180000395155:15004-15130 - | C---CTTGGAA------ACAGAAGTTGT---GGAAGAGAAGCGCAAAACAAAGAAGGTCAAGAAACCTAAGAAGAAGGTG---GTAA---------CGGAA-------GAGGAACAGGAAG---------AACAACTGGAAGAACAACCAGA---------------------------------------------AGA-------------------------------------------------------------------------------------------------------------------------------------GGTGG---------AAGAGC------------------------------------C | |
| dp5 | XR_group6:11993039-11993215 - | T---CGTAGAAGAACGAGCAGAGATCGT---GGAAGAAAAGAAAAAGGTGAAAAAGGTCAGGAAACCCAAGAGTAAGGTA---ACCG---------TAGAA-CAAGAAGCCGAAGAAGAAC---------AACCCGCAGAAGAGGTTGAAGA---------------------------------------------ACCA----------------------------------------------------------------------------------------------------------------------------------GAAGTGGAAGAACCCATCGCTGAAGA----ACCTCTTCCCGTAGAA------GAACCCA | |
| droPer2 | scaffold_33:11021-11152 - | C---TG--GAAGAGCAGGAGGATATCGT---AGAAGAAAAGAAAAAGGTCAAAAAGATCAAGAAACCAAAGAAAATTGTT---GAAA---------AGACT-------GAA---------------------------GAGGAGGTTCCTGAAGAGGAG---------------GTTCCCGA---------------AGAA----------------------------------------------------------------------------------------------------------------------------------GAGACTG---------TCGCAGA-----------------------------------A | |
| droWil2 | scf2_1100000004729:1310300-1310456 + | T---GATTGAGGATCAAGAAGAGATAGT---TGAAGAAAAACGCAAAATCAAGAAAGTTAAGAAACCAAAACAACCTAAG---GAGT---------TGACT----------GAAGAGGAAC---------AGCCCGAAGAGGAAATTCCAGA---------------------------------------------AGAG----------------------------------------------------------------------------------------------------------------------------------GAAGTCG---------TTGAGCA---AGAAGAAGTGGTCGA-AGAACAAG--------A | |
| droVir3 | scaffold_13049:12024708-12024843 + | A---TGTGGAAGAACAGGAAGAGATTGT---TGAAGAAAAACGTAAGATCAAGAAGGTTAAGAAACCCAAAATACCAGCTCTCGCTC---------AGAAA-------CAAATT---GAGC---------AATCTGAAGAGGAGGCTCCACA---------------------------------------------AGAA----------------------------------------------------------------------------------------------------------------------------------GAAACCC---------CTGAGGA------------------------------------ | |
| droMoj3 | scaffold_6680:8795943-8796091 - | T---TGAAGAAGAAGAGGAAGTAATAGT---GGAAGAAAAGCGTAAAATAAAGAAGGCAAAGAAACCAAAGAAAACTGTT---GAAGTAACTGAGGAGCCAG-------AAGAGCCGGAAG---------AGCCAACGGAAGACATTCCAGA---------------------------------------------AGAG----------------------------------------------------------------------------------------------------------------------------------GAAGTTG---------TCCAGGAGCAG-------------------------------- | |
| droGri2 | scaffold_15110:11465005-11465156 + | T---TGTTGAAGAGCAAGAAGATATTGT---TGAAGAAAAGCGTAAGGTAAAGAAAACAAAGAAACCCAAGAAGCCCGTA---GAA----------------------------CTGGAAC---------AGGCTACAGAAGACATTCCACA---------------------------------------------AGAA----------------------------------------------------------------------------------------------------------------------------------GAGCTCA---------TTGAAGAGCAGGAAGAAATTGTCGA-GGAG---AAGCG----C |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/17/2015 at 11:08 PM