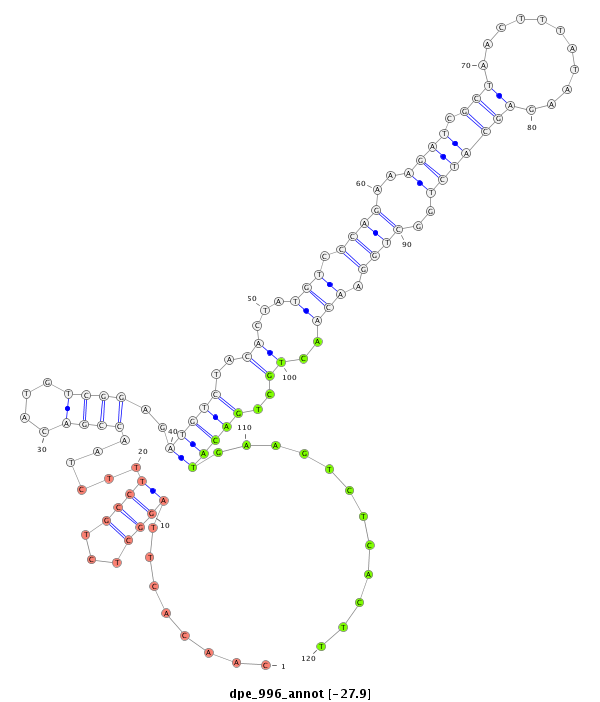
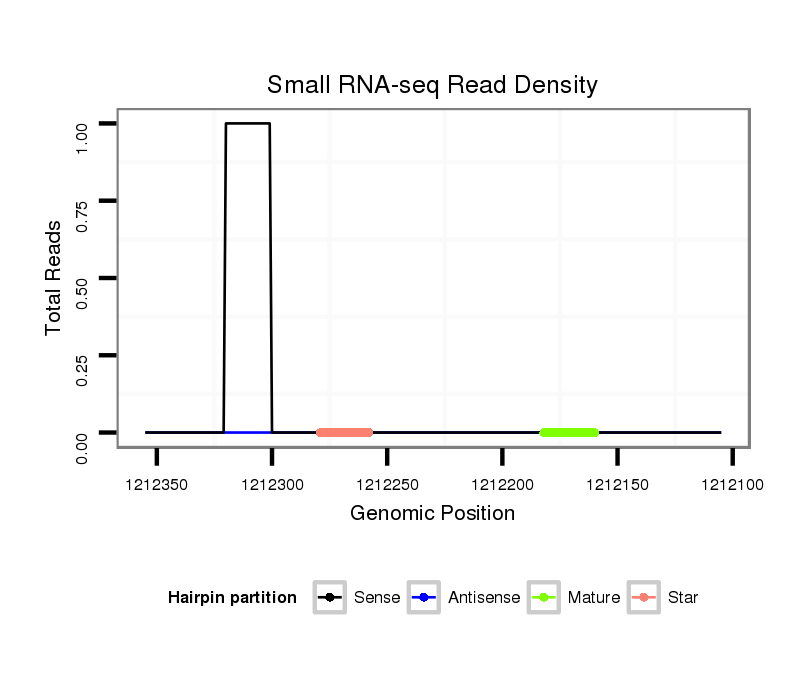
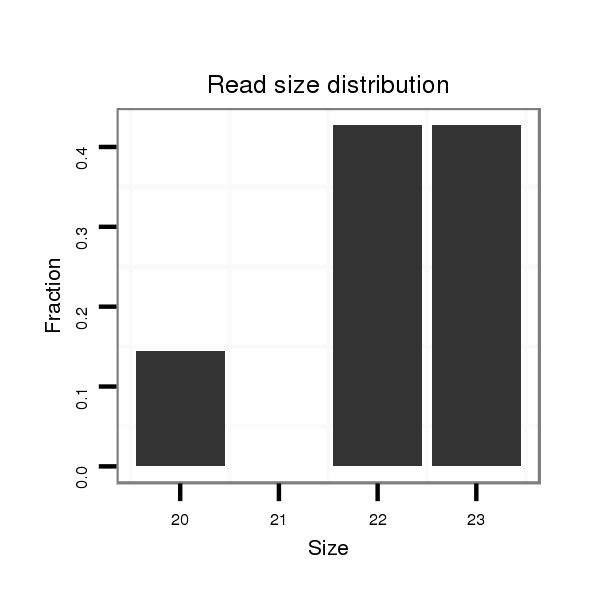
| TGCCGTAGCTGTAGAGACGAAGAAATGGAAGAAACGGTAGA-----GCATCTACTATGCCACCGTCCAGCACTGCAAGCAA-AAAGACTTGCCCAATTAGGAAGCAGATT-TCTGATAGACACGTCCGACCTGTCTGACACCAATCCACATCGGATACAAAAATTCACCAGCGCCTCTGGCTGGGAAAACTGGTAAC--------TTC-ACACGA---------AC---------CTCGACGG--GCAAAAGGGGAA----------------AACATACATGGTATCACAATGGACCTCTAATAGTCTAAGT | Size | Hit Count | Total Norm | Total | M026
Head | M043
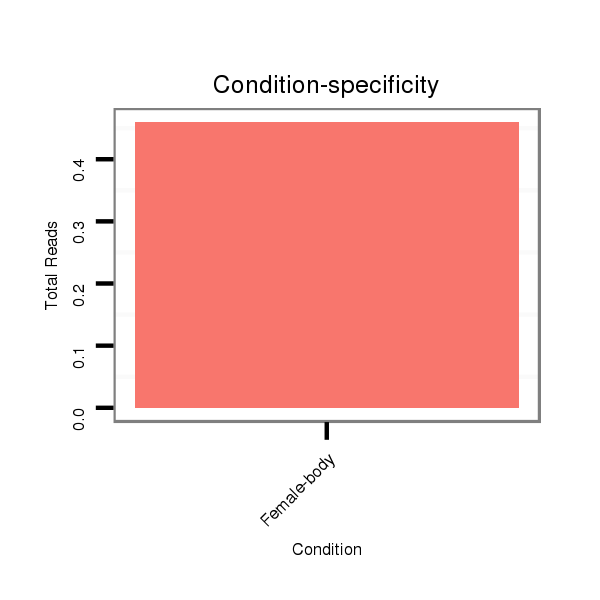
Female-body | M056
Embryo | V046
Embryo | V052
Head | V058
Head | V120
Male-body | SRR1275488
Male larvae | SRR1275486
Male prepupae | SRR1275484
Male prepupae | GSM1528802
follicle cells |
| ........................................................TGCCACCGTCCAGCACTGCAAGCAA-A...................................................................................................................................................................................................................................... | 26 | 1 | 5.00 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| ..............................................GCATCTACTATGCCACCGTCCAGCACT................................................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................TGGTAAC--------TTC-ACACGA---------AC---------CTCGACGG--GC.................................................................. | 28 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................TGGGAAAACTGGTAAC--------TTC-A....................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................CTGATAGACACGTCCGACCTGTCTGA............................................................................................................................................................................... | 26 | 3 | 0.67 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................ATT-TCTGATAGACACGTCCGACCTG.................................................................................................................................................................................... | 25 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................T-TCTGATAGACACGTCCGACCTGTC.................................................................................................................................................................................. | 25 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................GGTAAC--------TTC-ACACGA---------AC---------CTCGACGG--GCAT................................................................ | 29 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................TCCAGCACTGCAAGCAA-AAAGACT................................................................................................................................................................................................................................ | 24 | 4 | 0.50 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................ATGGAAGAAACGGTAGA-----GCATCTAC................................................................................................................................................................................................................................................................... | 25 | 20 | 0.50 | 10 | 0 | 2 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 5 |
| ................................................................................................................CTGATAGACACGTCCGACCTGTCTG................................................................................................................................................................................ | 25 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TCCACATCGGATACAAAAATTCACCAGC............................................................................................................................................. | 28 | 20 | 0.30 | 6 | 0 | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................ATGGAAGAAACGGTAGA-----GCAT....................................................................................................................................................................................................................................................................... | 21 | 20 | 0.15 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| .................................................................................................................................................................................................TAAC--------TTC-ACACGA---------AC---------CTCGACGG--G................................................................... | 24 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................ATGGAAGAAACGGTAGA-----GCATCT..................................................................................................................................................................................................................................................................... | 23 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................TAAC--------TTC-ACACGA---------AC---------CTCGACGG--GC.................................................................. | 25 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................TAAC--------TTC-ACACGA---------AC---------CTCGACGG--GCA................................................................. | 26 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................TCGGATACAAAAATTCACCAGCGCC.......................................................................................................................................... | 25 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................ATGGAAGAAACGGTAGA-----GCATCTA.................................................................................................................................................................................................................................................................... | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................ATGGAAGAAACGGTAGA-----GCATC...................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................ATACAAAAATTCACCAGCGCCTC........................................................................................................................................ | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TCCACATCGGATACAAAAATTCACCAG.............................................................................................................................................. | 27 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................TAAC--------TTC-ACACGA---------AC---------CTCGACG....................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................AAAAATTCACCAGCGCCTCTGGCTGG................................................................................................................................. | 26 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TCCACATCGGATACAAAAATTCACC................................................................................................................................................ | 25 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................AGAAATGGAAGAAACGGTAGA-----GCATC...................................................................................................................................................................................................................................................................... | 26 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................GGATACAAAAATTCACCAGCGCCTC........................................................................................................................................ | 25 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................TACAAAAATTCACCAGCGCCTCTGGC.................................................................................................................................... | 26 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................C-ACACGA---------AC---------CTCGACGG--GCAA................................................................ | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................ATAGACACGTCCGACCTG.................................................................................................................................................................................... | 18 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................AGAAATGGAAGAAACGGTAGA-----GCA........................................................................................................................................................................................................................................................................ | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |