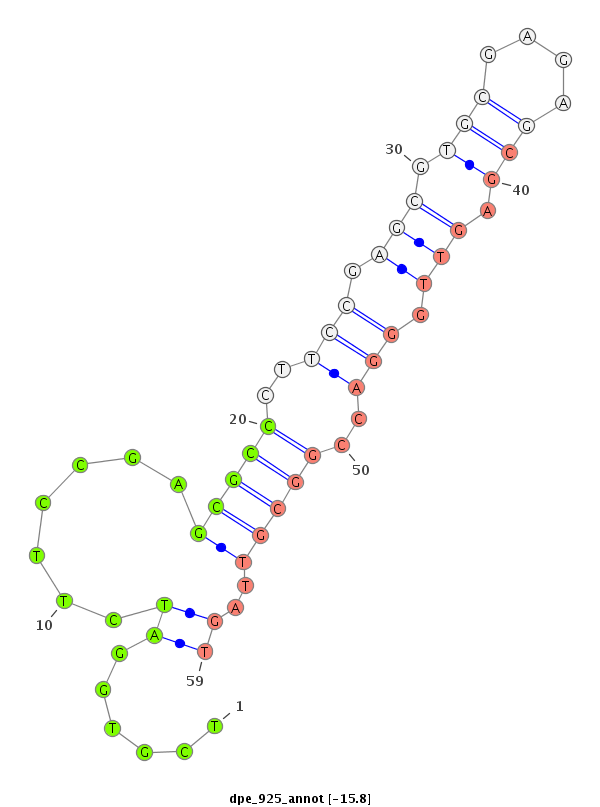
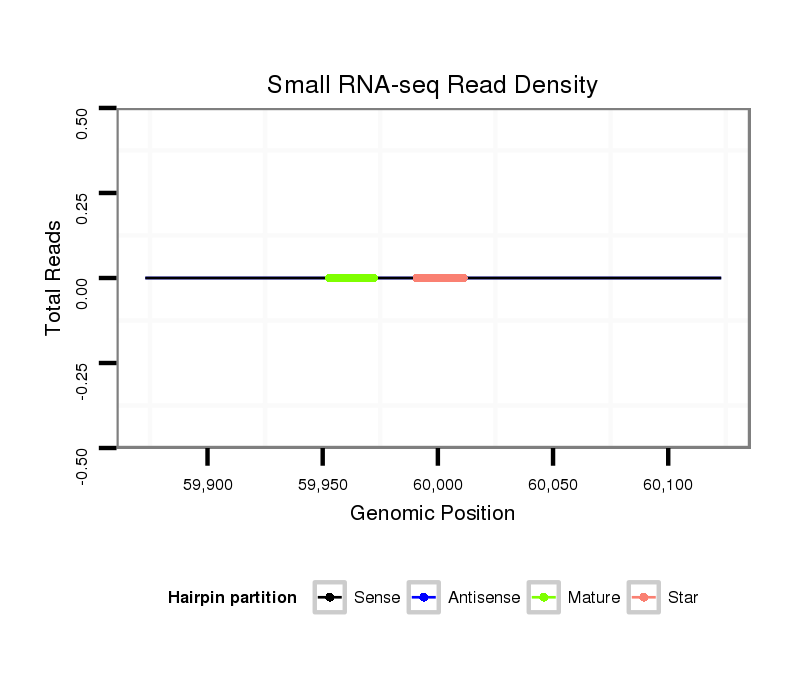
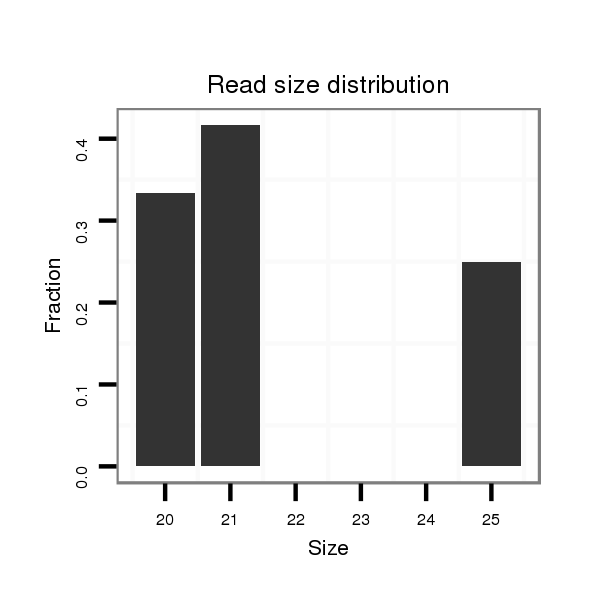
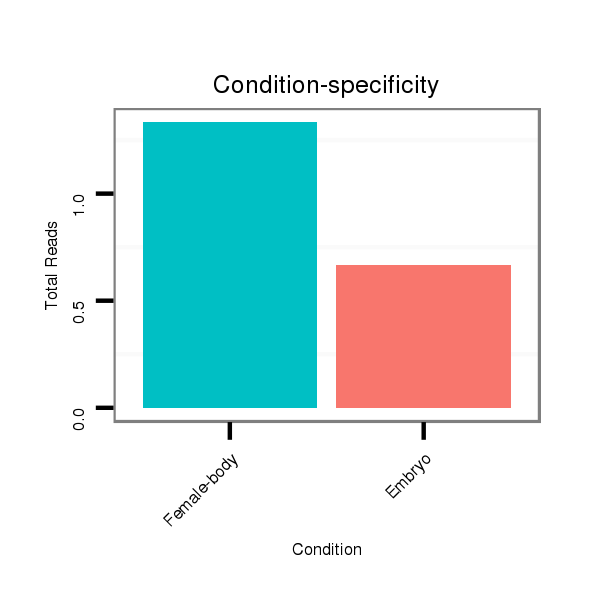
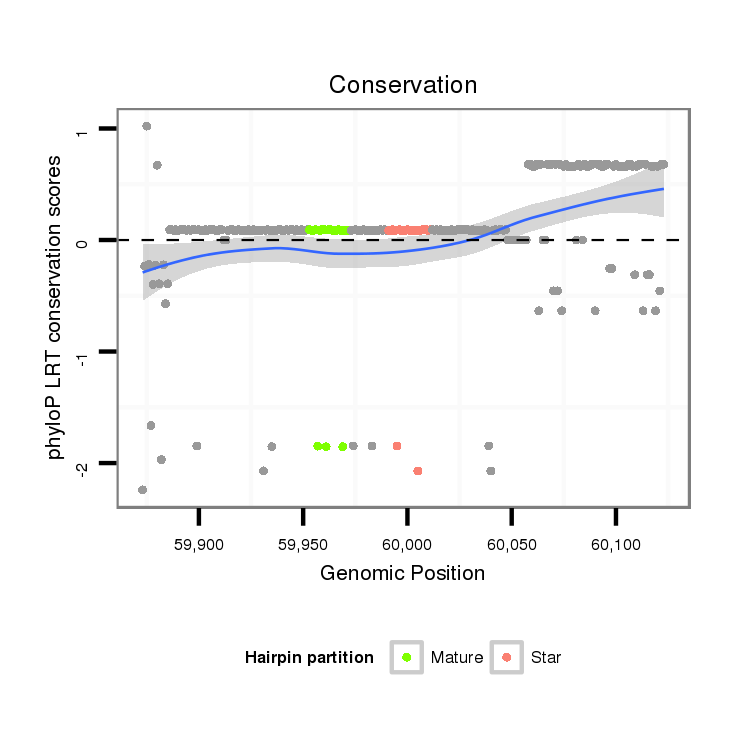
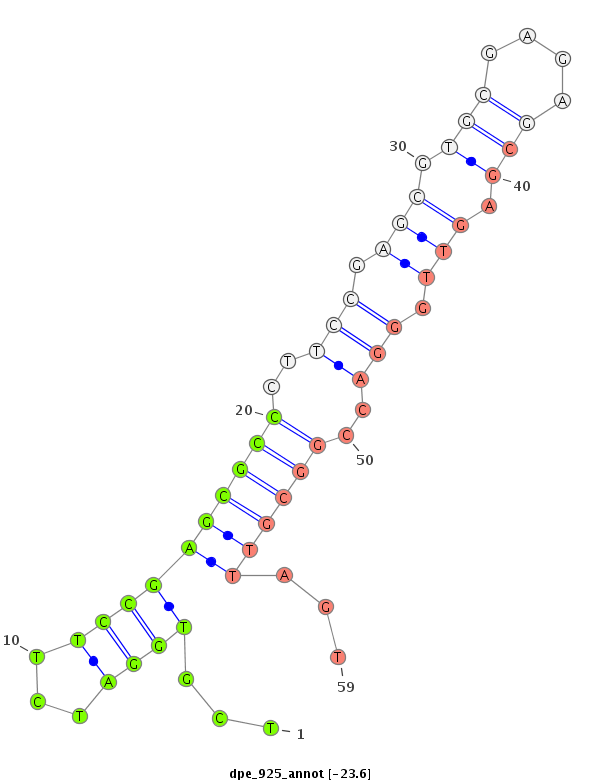
| TGAAAGCGTAACTAACTCTAAGTCTTGTGCTCTAAGCTT--GATGCTGCGGTGAGCGCCGTGTGGATCGTGTAGTGTCGATCGTAGATTTTCCGAGTGCCCCTCCGAGCGCGCGAGAGCGAGCTGGGACCGGAGTTAGTACAGTGCGCAGCAATTATCTGCGGCTGCACTGAACA---------------------------------------------------------------------------- | Size | Hit Count | Total Norm | Total | M022
Male-body | M040
Female-body | M059
Embryo | M062
Head | V043
Embryo | V051
Head | V112
Male-body | SRR902010
Ovary | SRR902011
Testis | SRR902012
CNS imaginal disc |
| .....GCGTAACTAACTCTAAGTCTTGTGCTC........................................................................................................................................................................................................................... | 27 | 3 | 1.00 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................TGGATCGTGTAGTGTCGATCGTAGATTT................................................................................................................................................................. | 28 | 4 | 0.50 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| ..............................................................TGGATCGTGTAGTGTCGATCGTAGA.................................................................................................................................................................... | 25 | 4 | 0.50 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| ..........................................................................TGTCGATCGTAGATTTTCCGAGTGC........................................................................................................................................................ | 25 | 4 | 0.50 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................TGGATCGTGTAGTGTCGATCGTAGATT.................................................................................................................................................................. | 27 | 4 | 0.50 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| .........................TGTGCTCTAAGCTT--GATGCTGCGG........................................................................................................................................................................................................ | 24 | 4 | 0.50 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................TGCGGTGAGCGCCGTGTGGATCG...................................................................................................................................................................................... | 23 | 20 | 0.35 | 7 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....GCGTAACTAACTCTAAGTCTTGTGC............................................................................................................................................................................................................................. | 25 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............TAACTCTAAGTCTTGTGCTC........................................................................................................................................................................................................................... | 20 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....GCGTAACTAACTCTAAGTCTTGTGCT............................................................................................................................................................................................................................ | 26 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....AGCGTAACTAACTCTAAGTCTTG................................................................................................................................................................................................................................ | 23 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................TCCGAGTGCCCCTCCGAGCGCGCGA........................................................................................................................................ | 25 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................GTGGATCGTGTAGTGTCGATCGTAGATT.................................................................................................................................................................. | 28 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TGAGCGCCGTGTGGATCGTGTAGTGT.............................................................................................................................................................................. | 26 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................TGTGGATCGTGTAGTGTCGAT.......................................................................................................................................................................... | 21 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................GTCGATCGTAGATTTTCCGAGTGCCC...................................................................................................................................................... | 26 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................AATTATCTGCGGCTGCACT................................................................................. | 19 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................GAGCGCCGTGTGGATCGTGTAGTGT.............................................................................................................................................................................. | 25 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................CGCGCGAGAGCGAGCTGGGACCG........................................................................................................................ | 23 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TGAGCGCCGTGTGGATCGTG.................................................................................................................................................................................... | 20 | 20 | 0.20 | 4 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| ..............................................TGCGGTGAGCGCCGTGTGGATCGTG.................................................................................................................................................................................... | 25 | 20 | 0.15 | 3 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................TCCGAGCGCGCGAGAGCGAG................................................................................................................................. | 20 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................TGCGGTGAGCGCCGTGTGGATCGT..................................................................................................................................................................................... | 24 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................TGCGGTGAGCGCCGTGTGGATC....................................................................................................................................................................................... | 22 | 20 | 0.10 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................GCGGTGAGCGCCGTGTGGATCG...................................................................................................................................................................................... | 22 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TGAGCGCCGTGTGGATCGT..................................................................................................................................................................................... | 19 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................TCCGAGCGCGCGAGAGCGAGC................................................................................................................................ | 21 | 19 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................TGCTGCGGTGAGCGCCGTGTGG.......................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................CGGTGAGCGCCGTGTGGATC....................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................TGCTGCGGTGAGCGCCGTGTGGAA........................................................................................................................................................................................ | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................TGCTGCGGTGAGCGCCGTG............................................................................................................................................................................................. | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................TGCTGCGGTGAGCGCCGTGTGGATA....................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................GAGCGCCGTGTGGATCGTG.................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................CTGCGGTGAGCGCCGTGTGG.......................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................CTCCGAGCGCGCGAGAGCGAG................................................................................................................................. | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................CTGCGGTGAGCGCCGTGTGGATC....................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................TGCGGTGAGCGCCGTGTGGAT........................................................................................................................................................................................ | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |