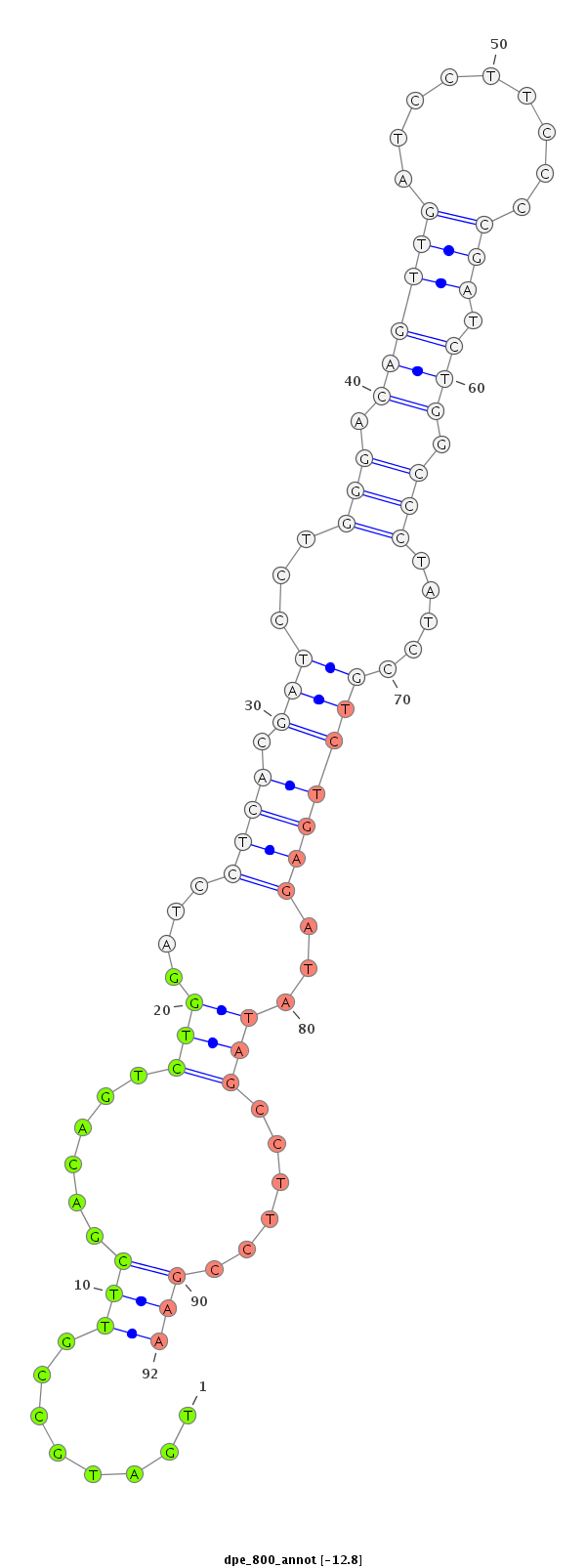
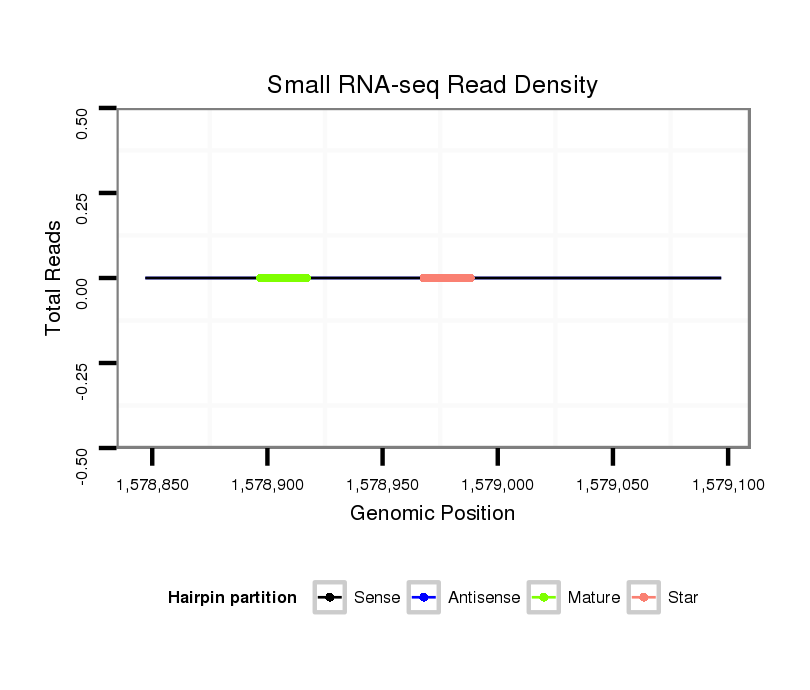
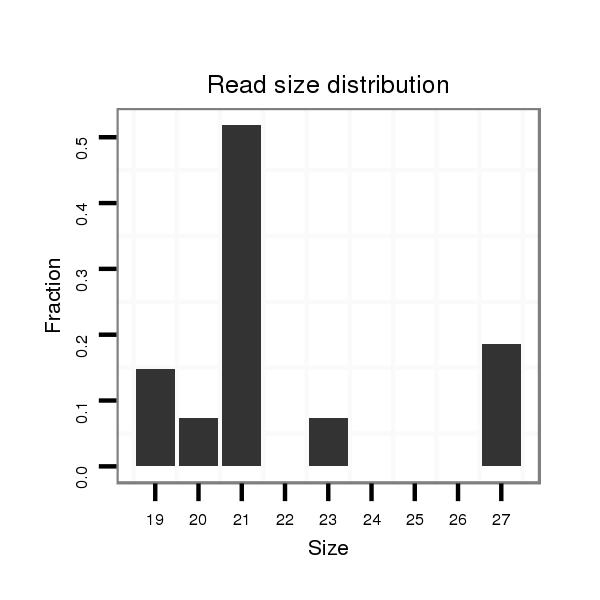
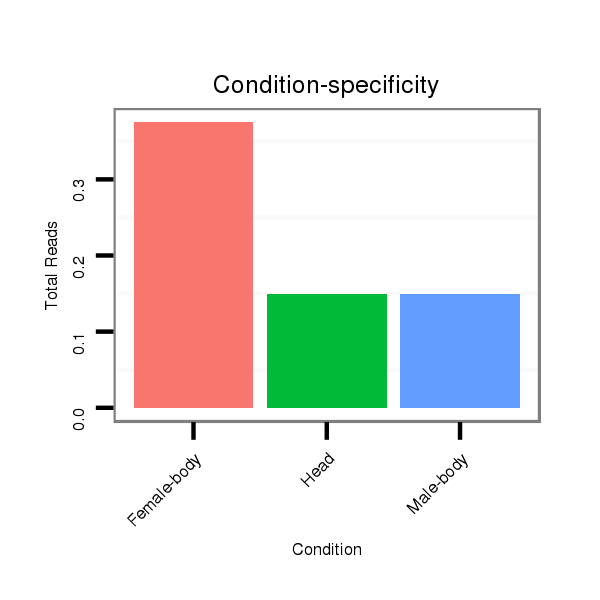
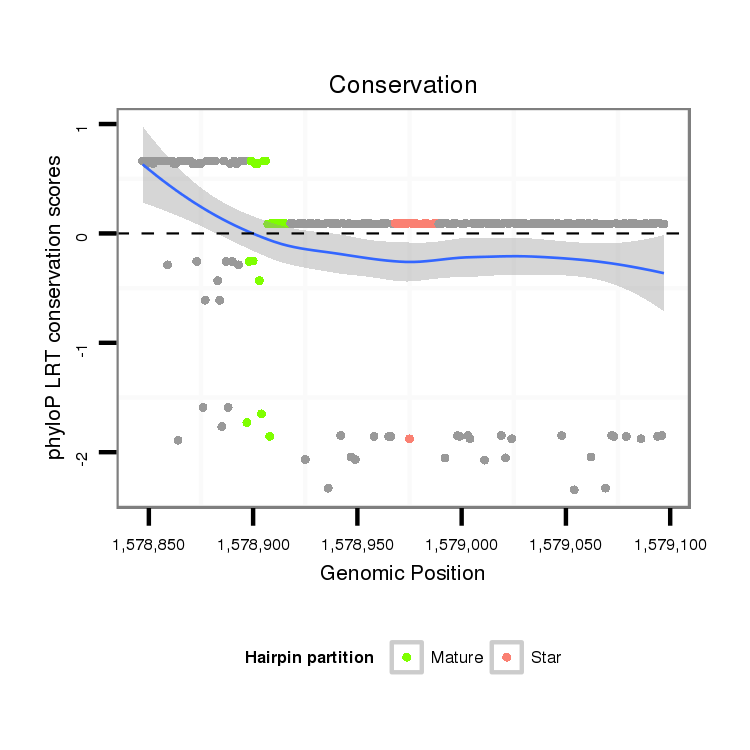
ID:dpe_800 |
Coordinate:scaffold_15:1578897-1579047 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
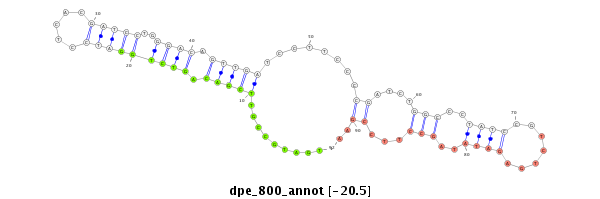
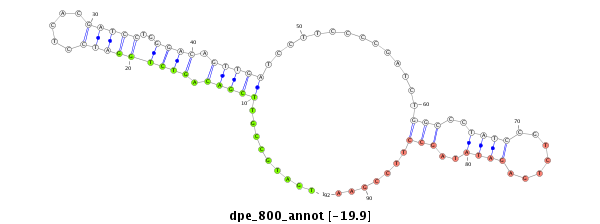
| -20.5 | -20.0 | -19.9 |
 |
 |
 |
CDS [Dper\GL16578-cds]; exon [dper_GLEANR_18225:2]; intron [Dper\GL16578-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TACATCATTAAATTTGCCAAAAATCGGCCTCATTTTCGAAGTCGAGATTTTGATGCCGTTCGACAGTCTGGATCCTCACGATCCTGGGACAGTTGATCCTTCCCCGATCTGGCCCTATCCGTCTGAGATATAGCCTTCCGAAGATGTGGATTCGCATTTCATTAAAATACTGATGTTTTCTGTACGCTATATCTCTTGCAGTCACTGGCTGTCCTTCTGCGGCCCACAGCCACGATTCCTCTGCTGTGTTG **************************************************........(((......(((....((((.(((...(((.((((((.........))).))).))).....)))))))...)))......)))************************************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | V057 head |
M042 female body |
V111 male body |
V042 embryo |
M021 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................................................CTTGTGCGGGCCACAGCCGCGA................ | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................TTTCGAAGTCGAGATTTTGATGCCGT................................................................................................................................................................................................ | 26 | 0 | 20 | 0.25 | 5 | 0 | 5 | 0 | 0 | 0 |
| ..................................................TGATGCCGTTCGACAGTCTGG.................................................................................................................................................................................... | 21 | 0 | 20 | 0.20 | 4 | 1 | 0 | 3 | 0 | 0 |
| .....................................................................................................................................................................................................................CTTGTGCGGCCCACAGCGGCG................. | 21 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................CCGTCTGAGTTATAACCGT.................................................................................................................. | 19 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................TCTGGATCCTCACGATCCTGGGACAGT.............................................................................................................................................................. | 27 | 0 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................GATTTTGATGCCGTTCGACA.......................................................................................................................................................................................... | 20 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 |
| .................................TTTCGAAGTCGAGATTTTGATGCCG................................................................................................................................................................................................. | 25 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| .................................TTTCGAAGTCGAGATTTTGATGCC.................................................................................................................................................................................................. | 24 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| .....................................GAAGTCGAGATTTTGATGCCGTTC.............................................................................................................................................................................................. | 24 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................ATGCCGTTCGACAGTCTGGAT.................................................................................................................................................................................. | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................AGATTTTGATGCCGTTCGA............................................................................................................................................................................................ | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................................TCGAAGTCGAGATTTTGATGCCGT................................................................................................................................................................................................ | 24 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................AGTCTGGATCCTCACGATCCTGG.................................................................................................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................GATGCCGTTCGACAGTCTG..................................................................................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................TTCGACAGTCTGGATCCTCAC............................................................................................................................................................................ | 21 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................AGATTTTGATGCCGTTCGAC........................................................................................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ................................TTTTCGAAGTCGAGATTTTGATGCCGT................................................................................................................................................................................................ | 27 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................CGTTCGACAGTCTGGATCC................................................................................................................................................................................ | 19 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................GATGCCGTTCGACAGTCTGG.................................................................................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................TCTGAGATATAGCCTTCCGAA............................................................................................................. | 21 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................ATGTTTTCTGTCCGCTTTCT........................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................ATTTTGATGCCGTTCGACAGT........................................................................................................................................................................................ | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
|
ATGTAGTAATTTAAACGGTTTTTAGCCGGAGTAAAAGCTTCAGCTCTAAAACTACGGCAAGCTGTCAGACCTAGGAGTGCTAGGACCCTGTCAACTAGGAAGGGGCTAGACCGGGATAGGCAGACTCTATATCGGAAGGCTTCTACACCTAAGCGTAAAGTAATTTTATGACTACAAAAGACATGCGATATAGAGAACGTCAGTGACCGACAGGAAGACGCCGGGTGTCGGTGCTAAGGAGACGACACAAC
*************************************************************************************************************........(((......(((....((((.(((...(((.((((((.........))).))).))).....)))))))...)))......)))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M042 female body |
V111 male body |
V057 head |
M021 embryo |
|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................................CAGAAAGAAGCCGGGCGTCGGTGC................. | 24 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................CGGGTGCCGGTGCTGAGGAG.......... | 20 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................CGGAATGCTTCGACACCTAAG.................................................................................................. | 21 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ...............................................AAAACTACGGCAAGCTGTCAGACCTA.................................................................................................................................................................................. | 26 | 0 | 20 | 0.25 | 5 | 5 | 0 | 0 | 0 |
| .....................................................................................................................AGGTAGACTCGATATCGG.................................................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 |
| ..................................................ACTACGGCAAGCTGTCAGACC.................................................................................................................................................................................... | 21 | 0 | 20 | 0.15 | 3 | 0 | 0 | 0 | 3 |
| ........................GCCGGAGTAAAAGCTTCAGCTCT............................................................................................................................................................................................................ | 23 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 |
| ....................................................TACGGCAAGCTGTCAGACCTAGGAGT............................................................................................................................................................................. | 26 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ............................................................................................................................CTCTATATCGGAATGCTTCGACACCT..................................................................................................... | 26 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .................................................AACTACGGCAAGCTGGCAGACCTA.................................................................................................................................................................................. | 24 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .........................................................CAAGCTGTCAGACCTAGGAGTGCTA......................................................................................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ....................................................TACGGCAAGCTGTCAGGCCTAGG................................................................................................................................................................................ | 23 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| .....................................................ATCGCAAGCTGTCAGACCTA.................................................................................................................................................................................. | 20 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| .................................................................................................GGAAGGGGCAAGACACGGAT...................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| .................................AAAGCTTCAGCTCTAAAACTACGGCAAG.............................................................................................................................................................................................. | 28 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droPer2 | scaffold_15:1578847-1579097 + | dpe_800 | TACATCATTAAATTTGCCAAAAATCGGCCTCATTTTCGAAGTCGAGATTTTGATGCCGTTCGACAGTCTGGATCCTCACGATCCTGGGACAGTTGATCCTTCCCCGATCTGGCCCTATCCGTCTGAGATATAGCCTTCCGAAGATGTGGATTCGCATTTCATTAAAATACTGATGTTTTCTGTACGCTATATCTCTTGCAGTCACTGGCTGTCCTTCTGCGGCCCACAGCCACGATTCCTCTGCTGTGTTG |
| dp5 | 4_group3:841804-842054 - | TACATCATTAAATTTGCAAAAAATCGGCCCCATTTTCGTAGCCGAGATTTCGATGCCATTCAACAGTCTGGATCCTCAAGATCCTGGGAGAGTTGGTCCTGCACCGATCTGACCCTATTTGTCTGAGAAATAGCCTTCCGAAGATTTGGATCTGCACATCATTACAATACTGGTTTTATCTGTACGCTATATCTCTTGCAGCCACTGCCTGTCCTGCTGCGGGCCGTAGCCATGATTCCACTGCTGTATCG | |
| droEle1 | scf7180000491249:831980-832039 + | TACATCATTAAAATTGCAAAAAATCGACCCGATTTTACAAACCAAGTTTTTAACGCAGTT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
Generated: 06/08/2015 at 03:15 PM