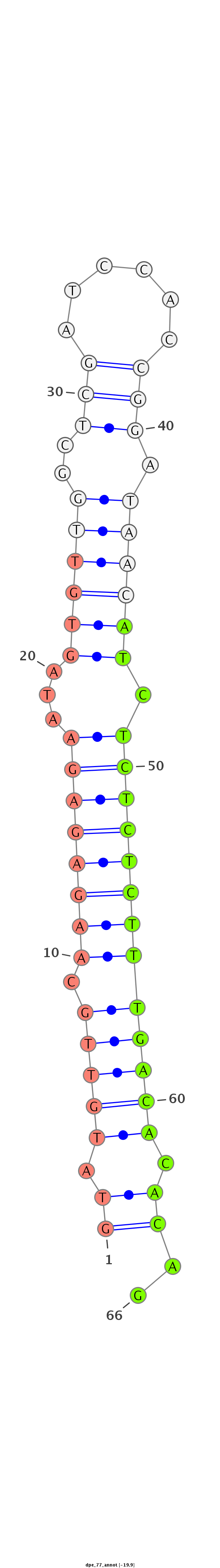
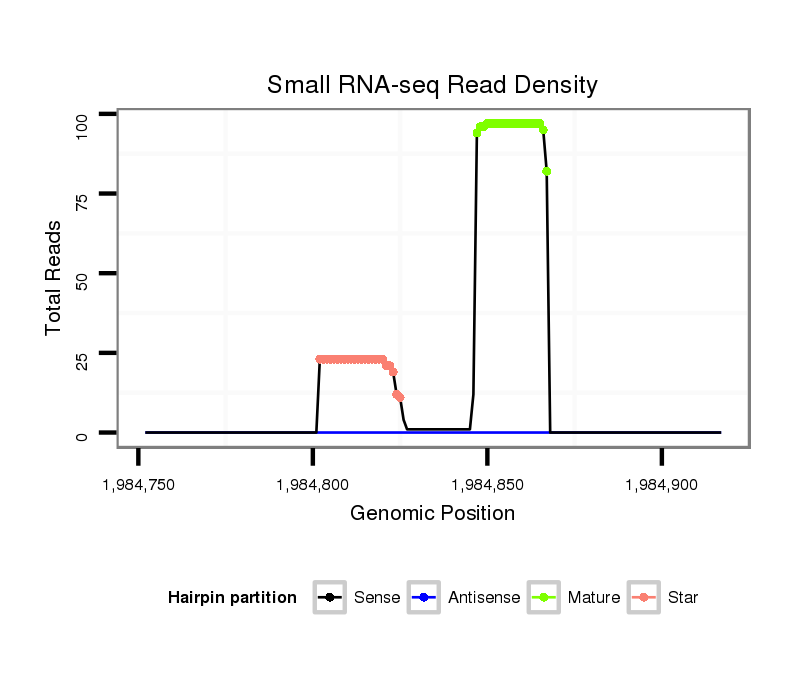
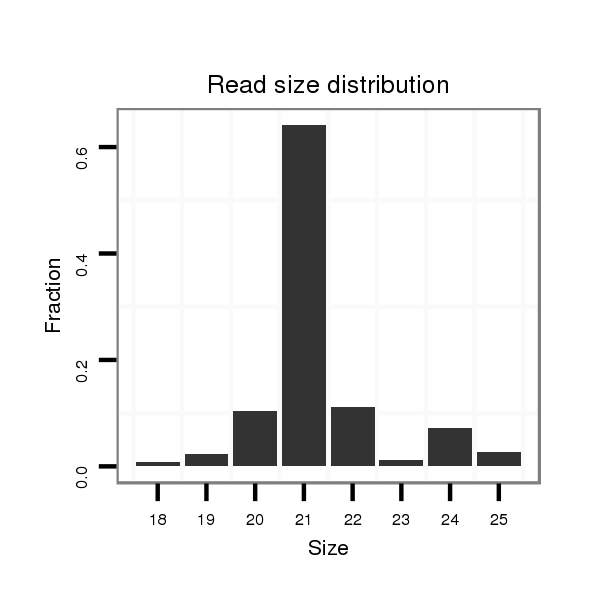
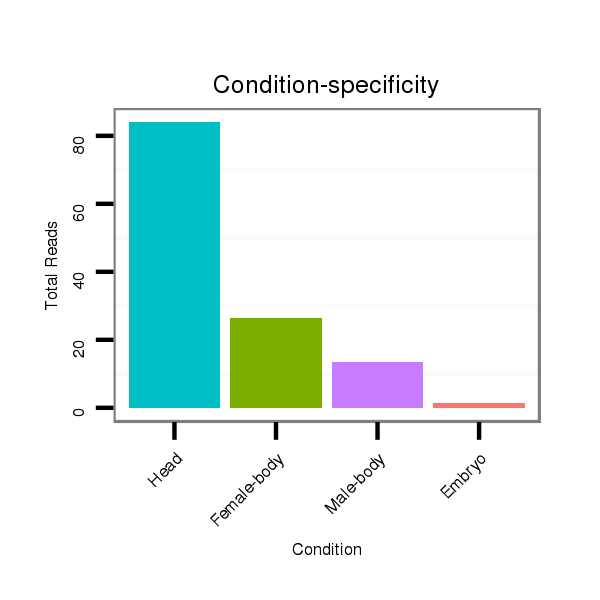
| ...............................................................................................ATCTCTCTCTTTGACACACAGT................................................. |
22 |
1 |
2 |
124.00 |
248 |
137 |
104 |
5 |
0 |
2 |
0 |
| ...............................................................................................ATCTCTCTCTTTGACACACAG.................................................. |
21 |
0 |
2 |
73.50 |
147 |
43 |
60 |
42 |
0 |
2 |
0 |
| ...............................................................................................ATCTCTCTCTTTGACACACA................................................... |
20 |
0 |
2 |
9.00 |
18 |
6 |
9 |
3 |
0 |
0 |
0 |
| ..................................................GTATGTTGCAAGAGAGAATAGTGT............................................................................................ |
24 |
0 |
2 |
8.50 |
17 |
7 |
0 |
0 |
10 |
0 |
0 |
| ..................................................GTATGTTGCAAGAGAGAATAGT.............................................................................................. |
22 |
0 |
2 |
7.00 |
14 |
0 |
2 |
0 |
12 |
0 |
0 |
| ..............................................................................................CATCTCTCTCTTTGACACACAG.................................................. |
22 |
0 |
2 |
6.50 |
13 |
5 |
6 |
2 |
0 |
0 |
0 |
| ..............................................................................................CATCTCTCTCTTTGACACACA................................................... |
21 |
0 |
2 |
4.00 |
8 |
2 |
5 |
0 |
0 |
0 |
1 |
| ..................................................GTATGTTGCAAGAGAGAATAGTGTT........................................................................................... |
25 |
0 |
2 |
3.50 |
7 |
4 |
1 |
0 |
2 |
0 |
0 |
| ................................................................................................TCTCTCTCTTTGACACACAG.................................................. |
20 |
0 |
2 |
2.50 |
5 |
0 |
0 |
5 |
0 |
0 |
0 |
| ................................................................................................TCTCTCTCTTTGACACACAGT................................................. |
21 |
1 |
2 |
2.50 |
5 |
4 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GTATGTTGCAAGAGAGAATAG............................................................................................... |
21 |
0 |
2 |
2.00 |
4 |
2 |
0 |
0 |
2 |
0 |
0 |
| ..................................................GTATGTTGCAAGAGAGAAT................................................................................................. |
19 |
0 |
2 |
2.00 |
4 |
2 |
2 |
0 |
0 |
0 |
0 |
| ..................................................GTATGTTGCAAGAGAGAATAGTG............................................................................................. |
23 |
0 |
2 |
1.50 |
3 |
1 |
1 |
0 |
1 |
0 |
0 |
| ..............................................................................................CATCTCTCTCTTTGACACAC.................................................... |
20 |
0 |
2 |
1.50 |
3 |
2 |
0 |
1 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTTTACACACAG.................................................. |
21 |
1 |
2 |
1.50 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTTTTTGACACACAG.................................................. |
21 |
1 |
2 |
1.50 |
3 |
0 |
1 |
2 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTTGCCACACAGT................................................. |
22 |
2 |
2 |
1.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................CATCTCTCTCTTTGACACACAGT................................................. |
23 |
1 |
2 |
1.00 |
2 |
0 |
1 |
1 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTTGACACAC.................................................... |
19 |
0 |
2 |
1.00 |
2 |
0 |
2 |
0 |
0 |
0 |
0 |
| ..........................................................................TGGCTCGATCCACCGGATAAC....................................................................... |
21 |
0 |
2 |
1.00 |
2 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTTGACACACAGTT................................................ |
23 |
2 |
2 |
1.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTATCTCTTTGACACACAG.................................................. |
21 |
1 |
2 |
1.00 |
2 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GTATGTTGCAAGAGAGAATAGC.............................................................................................. |
22 |
1 |
2 |
1.00 |
2 |
1 |
0 |
0 |
1 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTTCACACACAG.................................................. |
21 |
1 |
2 |
1.00 |
2 |
0 |
2 |
0 |
0 |
0 |
0 |
| ..................................................................................................TCTCTCTTTGACACACAGT................................................. |
19 |
1 |
2 |
1.00 |
2 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................................................................TCTCTCTTTGACACACAG.................................................. |
18 |
0 |
2 |
1.00 |
2 |
1 |
1 |
0 |
0 |
0 |
0 |
| ................................................................................................TCTCTCTCTTTGACACACAGTT................................................ |
22 |
2 |
2 |
1.00 |
2 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCACTCTTTGACACACAGT................................................. |
22 |
2 |
2 |
1.00 |
2 |
0 |
2 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCGTTGACACACAGT................................................. |
22 |
2 |
2 |
1.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCCCTTTGACACACAGT................................................. |
22 |
2 |
2 |
1.00 |
2 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................................................AGCTCTCTCTTTGACACACAGT................................................. |
22 |
2 |
2 |
1.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTTGACACACAT.................................................. |
21 |
1 |
2 |
1.00 |
2 |
1 |
0 |
1 |
0 |
0 |
0 |
| ...............................................................................................ATATCGCTCTTTGACACACAG.................................................. |
21 |
2 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTGGACACACAGT................................................. |
22 |
2 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTATCTCTTTGACACACAGT................................................. |
22 |
2 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..................................................GTATGTTGCGAGAGTGAATAGTC............................................................................................. |
23 |
3 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
| ...............................................................................................ATCTCTCTCTTTGTCACACAGT................................................. |
22 |
2 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATGTCTCTCTTTGACACACAG.................................................. |
21 |
1 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
| ..................................................................................................TCTCTCTTTGACACACCG.................................................. |
18 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATATCTCTCTTTGACACACAGT................................................. |
22 |
2 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...................................................TATGTTGCAAGAGAGAATAGTGTT........................................................................................... |
24 |
0 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCGCTTTGAAACACAG.................................................. |
21 |
2 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTTGACACCCAGT................................................. |
22 |
2 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..................................................GTATGTTGCAAGAGAGAATAA............................................................................................... |
21 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..................................................GTATGTTGCAAGAGACAATTCTGTT........................................................................................... |
25 |
3 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCCCTCTTTGACACACAGT................................................. |
22 |
2 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ACCTCTCTCTTTGACACACAGT................................................. |
22 |
2 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ................................................................................................TATCTCTCTTTGACACACAGC................................................. |
21 |
2 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCCCTCTCCTTGACACACAGT................................................. |
22 |
3 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTCGACACACAGT................................................. |
22 |
2 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTGTGACACACAGT................................................. |
22 |
2 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCGCTCTCTTTGACACACAG.................................................. |
21 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..................................................GTATGTTGCAAGAGAGAATAGAG............................................................................................. |
23 |
1 |
2 |
0.50 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
| ..............................................................................................CATCTCTCACTTTGACACACA................................................... |
21 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTTGACACACAGC................................................. |
22 |
1 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ..................GCAGAACTACACCGAGCC.................................................................................................................................. |
18 |
0 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
| ..............................................................................................CATCTCTCCCTTTGACACACAG.................................................. |
22 |
1 |
2 |
0.50 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTTGACACGCAGT................................................. |
22 |
2 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................AGCGCTCTCTTTGACACACAG.................................................. |
21 |
2 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTTGACCCACAGT................................................. |
22 |
2 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTTGACACACAA.................................................. |
21 |
1 |
2 |
0.50 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| ...............................................................................................ATCTTTCTCTTTGACACACAGT................................................. |
22 |
2 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTTGACACACAGA................................................. |
22 |
1 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ....................................................ATGTTGCAAGAGAGAATAGTGT............................................................................................ |
22 |
0 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCGCTCTTTGACACACAG.................................................. |
21 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..................................................GTATGGTGCAAGAGAGAATAGTGTT........................................................................................... |
25 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCCTTGACACACAG.................................................. |
21 |
1 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTTGACACACAGG................................................. |
22 |
0 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTTGCCACACAG.................................................. |
21 |
1 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCCCTCTCTTTGACACACAGT................................................. |
22 |
2 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTTGACACACCGT................................................. |
22 |
2 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTTTACACACAGT................................................. |
22 |
2 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................TTCTCTCTCTTTGACACACAGT................................................. |
22 |
2 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..................................................GTATGTTGCAAGAGAGAATAGTGA............................................................................................ |
24 |
1 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................................................CTCTCTCTCTTTGACACACAG.................................................. |
21 |
1 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTATCTCTTGGACACACAG.................................................. |
21 |
2 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTATCTCTTTGACACACTGT................................................. |
22 |
3 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................TCTCGCTCTTTGACACAC.................................................... |
18 |
1 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..................................................GTATGCTGCAAGTGAGAATACT.............................................................................................. |
22 |
3 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATATCTCTCATTGACACACAGC................................................. |
22 |
3 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCATTGACACACAGTT................................................ |
23 |
3 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTAGCGCTTTGACACACAGG................................................. |
22 |
3 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTATATCTTTGACACACAGT................................................. |
22 |
3 |
6 |
0.33 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTATCTCTTTGCCACACAGT................................................. |
22 |
3 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCGCTCTTTGACACTCAGT................................................. |
22 |
3 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTCTGACACACGG.................................................. |
21 |
2 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATATCCCTCTTTGACAAACAG.................................................. |
21 |
3 |
5 |
0.20 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTTGACCAACAGT................................................. |
22 |
3 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATGTGTCTCTTTGACACACAGT................................................. |
22 |
3 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTTGACACACACT................................................. |
22 |
2 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTTTTTTACACACAGT................................................. |
22 |
3 |
6 |
0.17 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTTGCCACACAGTC................................................ |
23 |
3 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTTTCCACACAGT................................................. |
22 |
3 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| .........CAAGGACGAGCAGATGTGCAC........................................................................................................................................ |
21 |
3 |
7 |
0.14 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTTCACACACAGT................................................. |
22 |
2 |
7 |
0.14 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATATCTCTCTTTCACACACAGT................................................. |
22 |
3 |
13 |
0.08 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................ATCTCTCTCTTTTACCCACAT.................................................. |
21 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |