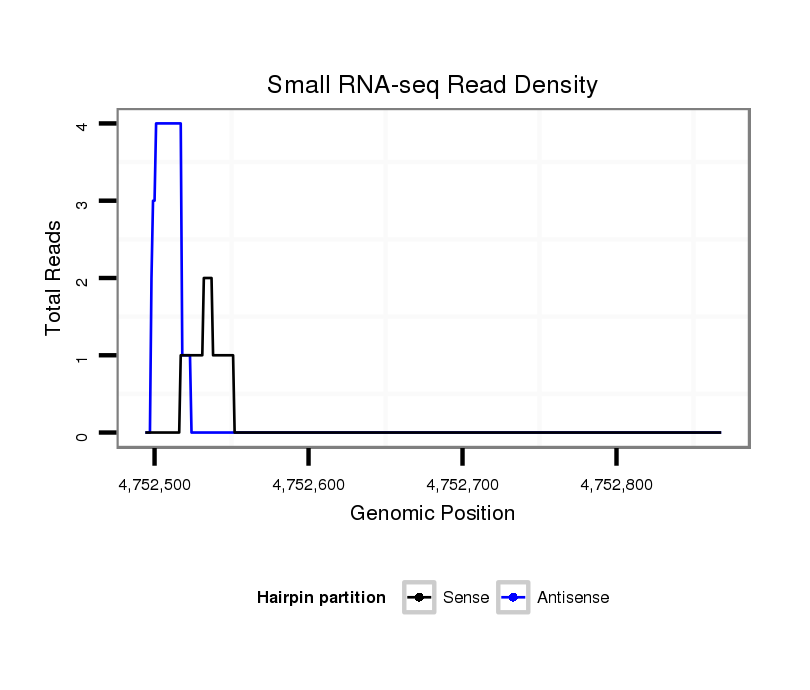
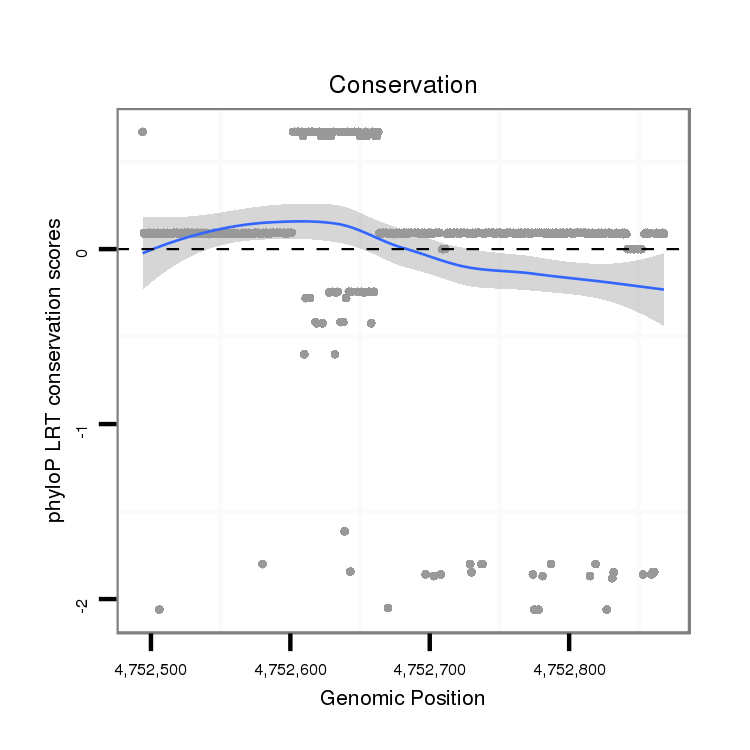
ID:dpe_562 |
Coordinate:scaffold_1:4752544-4752818 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dper\GL19350-cds]; exon [dper_GLEANR_21490:2]; exon [dper_GLEANR_21490:1]; CDS [Dper\GL19350-cds]; intron [Dper\GL19350-in]
No Repeatable elements found
| ##################################################-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------#################################----------------- TTCGACACTCCACCGAGCACCGTAAAGCTATTTTGTTGGAACTGTCACCGGTACGTGTCCGTCGCAGAGGCGTCAGTCGCTTTTGCTTTTGGTTAAAAAATTTGAAATTTATTTTGCTAAATTTTGTTCCAACTCGAACGAAATACTTTGAATTTTGAGGTGTTGTTCGTACAAAATGTGGGAGTGGAATGAAATTTTTGCTACGAAAGATTATCGTCGTTTGTTTTCATAAATCTAAGATTTTTGAAAATATCAAATGCCAAAAAAGCCTTTGATTTCTCCATCTAATCTAGTAGTTTCTTAGTTATTCTTATTATTTGTACAGTCTGTGGACTTCTGGAGGCAGTCACAAATCTAGCCCTTCTCAAGACCCAC **************************************************(((((...(((..(((((((((.....)))))))))...))).............((((((......))))))((((((......))))))((((((((((...))))))))))...)))))....((((.(((((((((((.......(((((...((((((.........(((((((.(((((...))))).)))))))((((((.((.......)).)))))).........)))))).))))).......)))))))....)))).)))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V111 male body |
V050 head |
V057 head |
|---|---|---|---|---|---|---|---|---|
| .......................AAAGCTATTTTGTTGGAACTG........................................................................................................................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ......................................GAACTGTCACCGGTACGTGT............................................................................................................................................................................................................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .......................................................................................................................................................................CGTACAAATTATGGGAGTGTAA.......................................................................................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................TGCTTAGTTATTCTTATT............................................................ | 18 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 |
| .......................................................................................................................................................................CGTACAAATTATGGGAGTGTA........................................................................................................................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 |
|
AAGCTGTGAGGTGGCTCGTGGCATTTCGATAAAACAACCTTGACAGTGGCCATGCACAGGCAGCGTCTCCGCAGTCAGCGAAAACGAAAACCAATTTTTTAAACTTTAAATAAAACGATTTAAAACAAGGTTGAGCTTGCTTTATGAAACTTAAAACTCCACAACAAGCATGTTTTACACCCTCACCTTACTTTAAAAACGATGCTTTCTAATAGCAGCAAACAAAAGTATTTAGATTCTAAAAACTTTTATAGTTTACGGTTTTTTCGGAAACTAAAGAGGTAGATTAGATCATCAAAGAATCAATAAGAATAATAAACATGTCAGACACCTGAAGACCTCCGTCAGTGTTTAGATCGGGAAGAGTTCTGGGTG
**************************************************(((((...(((..(((((((((.....)))))))))...))).............((((((......))))))((((((......))))))((((((((((...))))))))))...)))))....((((.(((((((((((.......(((((...((((((.........(((((((.(((((...))))).)))))))((((((.((.......)).)))))).........)))))).))))).......)))))))....)))).)))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V111 male body |
V057 head |
V050 head |
M042 female body |
V042 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| ....TGTGAGGTGGCTCGTGGCAT............................................................................................................................................................................................................................................................................................................................................................... | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .......GAGGTGGCTCGTGGCATTTCGAT......................................................................................................................................................................................................................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....GTGAGGTGGCTCGTGGCAT............................................................................................................................................................................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....AAAGAGGTGGCTCGTGGCATTTCG........................................................................................................................................................................................................................................................................................................................................................... | 24 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................TGGCCAAGCACAGCCAGCGTTT................................................................................................................................................................................................................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................................TGGGCAAGCACAGCCAGCGTCT................................................................................................................................................................................................................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................AGCTTCACAGTGGGCATGC................................................................................................................................................................................................................................................................................................................................ | 19 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................ACGTTGACAGTGGGCATGTA............................................................................................................................................................................................................................................................................................................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................GTTTCTGAAACTAAAGAG.............................................................................................. | 18 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................................................................................................................................CGAAAAGTAAAGAGGCAGAT........................................................................................ | 20 | 3 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................ACCTTGACACTTGCCATCC................................................................................................................................................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................AGATCATCCAATAATGAAT.................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .........GGTCGGTCGTGGCATTCCG........................................................................................................................................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droPer2 | scaffold_1:4752494-4752868 + | dpe_562 | TTCGACACTCCACCGAGCACCGTAAAGCTATTTTGTTGGAACTGTCACCGGTACGTGTCCGTCGCAGAGGCGTCAGTCGCTTTTGCTTTTGGTTAAAAAATTTGAAATTTATTTTGCTAAATTTTGTTCCAACTCGAACGAAATACTTTGAATTTTGAGGTGTTGTTCGTACAAAATGTGGGAGTGGAATGAAATTTTTGCTACGAAAGATTATCGTCGTTTGTTTTCATAAATCTAAGATTTT---TGAAAATATCAAATGCCAAAAAAGCCTTTGATTTCTCCATCTAATCTAGTAGTTTCTTAGTTATTCTTATTATTTGTACAGTCTGTG-----------------------GACTTCTGGAGGCAGTCACAAATCTAGCCCTTCTCAAGACCCAC |
| dp5 | 4_group3:3271012-3271398 + | TTCGACACTCCAACGAGCACCGTAAAGCTATTTTGTTGGAACTGTCACCGGTACGTGTCCGTCGCAGAGGCGTCAGTCGCTTTTGCCTTTGGTTAAAAAATTTGAAATTTATTTTGCTAAATTTTGTTCCAACTCGAACGAAATATTTTTAATTTTGAGGTGTTGTTCGTACAAAAGGTGGGAGTGGAATGAAATTTTTGCTATGAAAGTTTATT---GTTTGTTTTCATAAATCCGAGATTTCGAACGAAAATATCAAATGCCAAAAAAGCCTTTGATTTCTTAATATATTCTAGCAGTTTCTTAGTTATTCTTATTATTTGTTCAGCCTGTGATTGTTACCTCCACAGTGGAAGCGAATTCAAGAGGCAGTC-----------TCTTCTTGGGACCCAC | |
| droBip1 | scf7180000396580:683217-683279 - | T-----------------------------------------------------------------------------------------------------------TTATTTTGGAAATTTTGTTTCAAACTTGGAGAGACTCCATCGGATTCTGGGATGCCTTCCGT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
Generated: 06/08/2015 at 02:54 PM