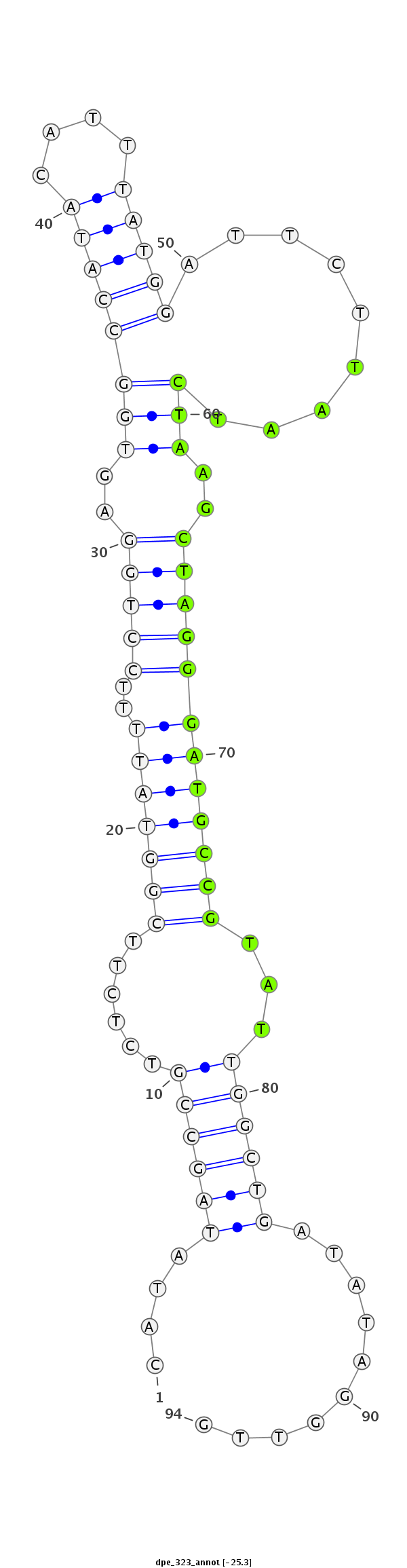
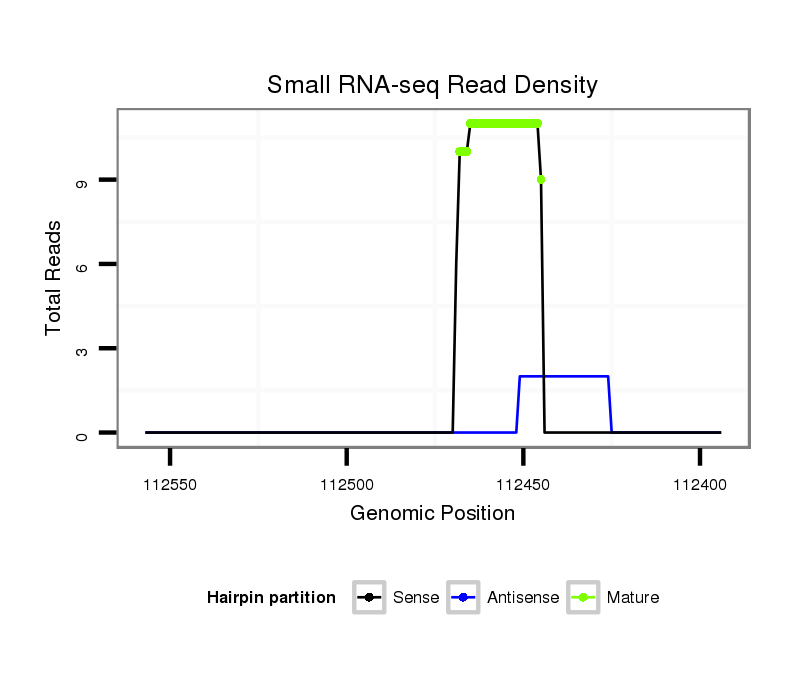
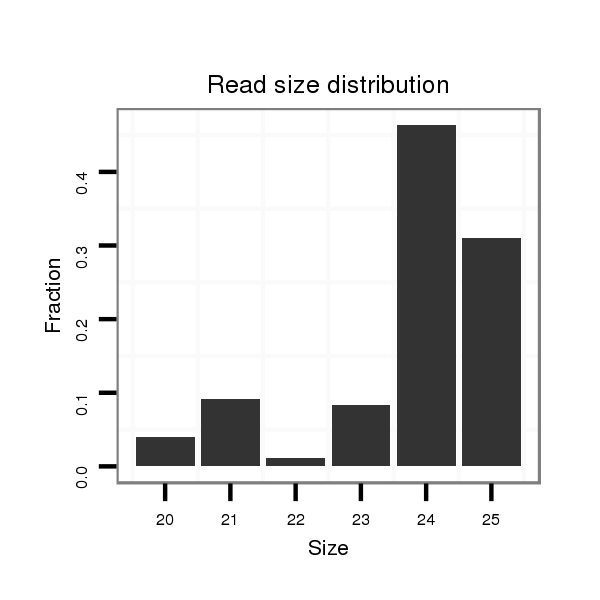
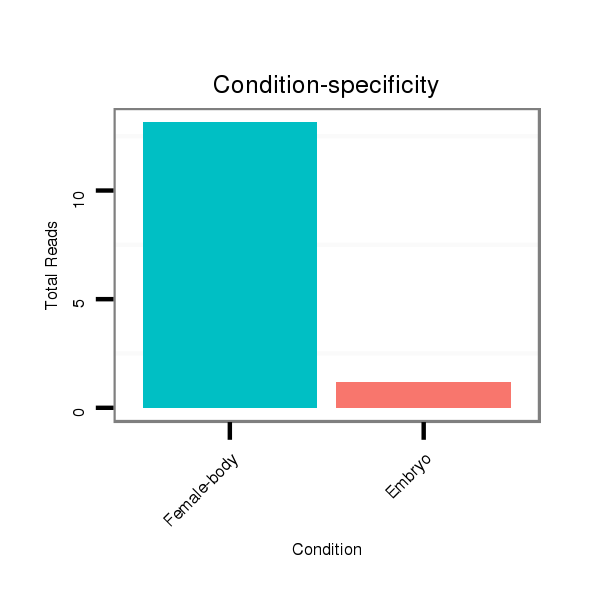
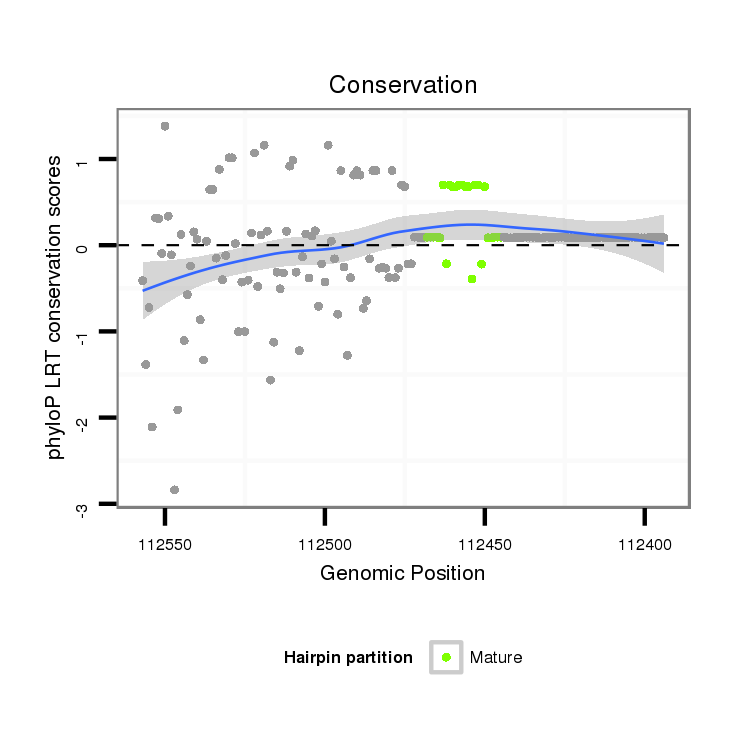
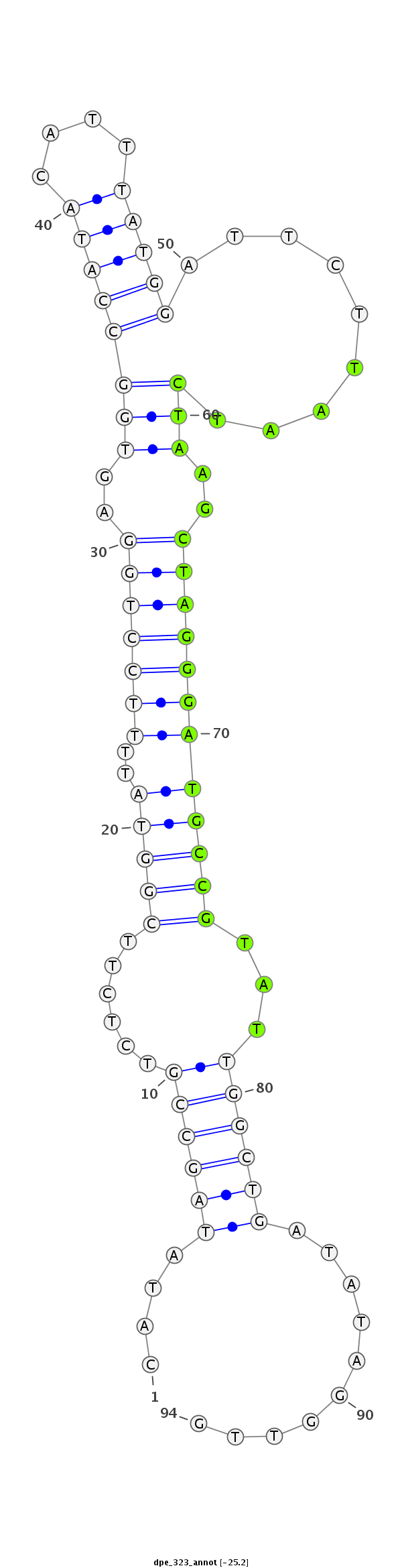
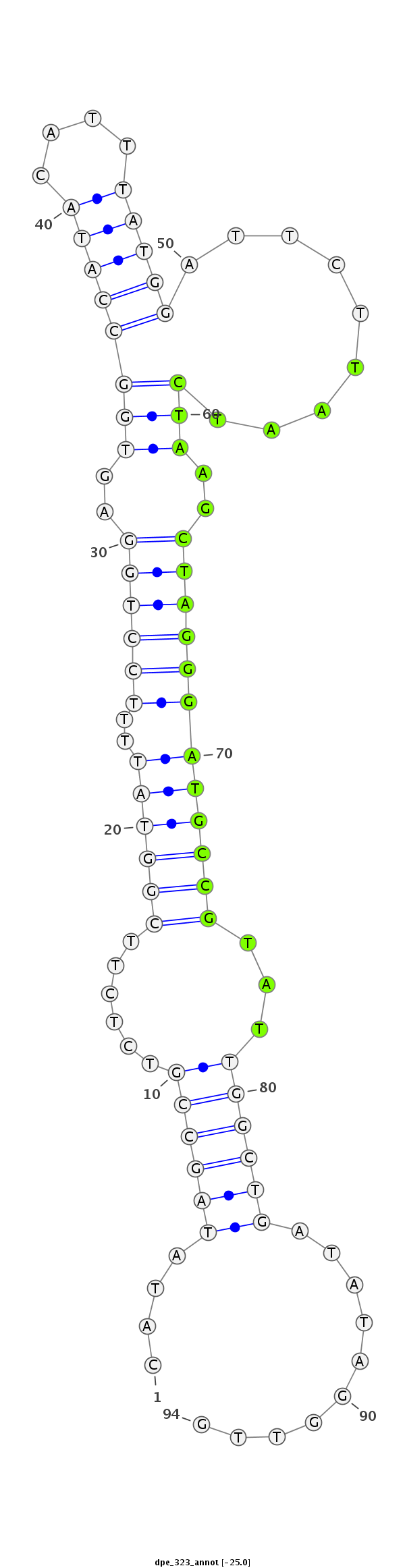
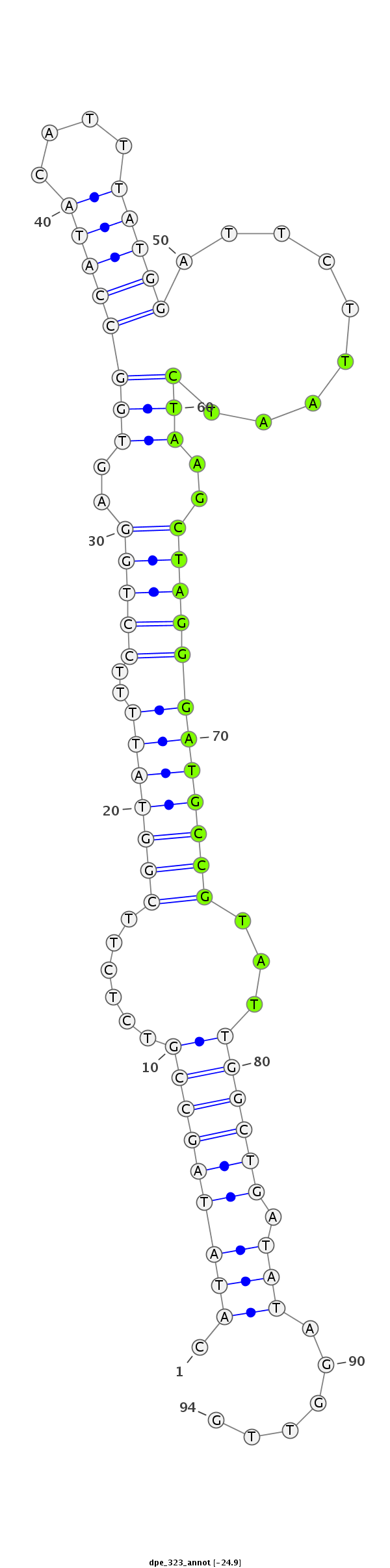
| .........................................................................................TAATCTAAGCTAGGGATGCCGTAT................................................... |
24 |
0 |
5 |
4.20 |
21 |
21 |
0 |
0 |
0 |
| ........................................................................................TTAATCTAAGCTAGGGATGCCGTAT................................................... |
25 |
0 |
5 |
4.00 |
20 |
18 |
0 |
2 |
0 |
| ........................................................................................TTAATCTAAGCTAGGGATGCCGTA.................................................... |
24 |
0 |
5 |
2.20 |
11 |
11 |
0 |
0 |
0 |
| ..........................................................................................................................TAGGTTGTAATGGGACTGCCGTAC.................. |
24 |
1 |
7 |
1.29 |
9 |
9 |
0 |
0 |
0 |
| ............................................................................................TCTAAGCTAGGGATGCCGTAT................................................... |
21 |
0 |
5 |
1.00 |
5 |
1 |
0 |
0 |
4 |
| ........................................................................................TTAATCTAAGCTAGGGACGCCGTAT................................................... |
25 |
1 |
6 |
0.83 |
5 |
5 |
0 |
0 |
0 |
| .........................................................................................TAATCTAAGCTAGGGACGCCGTAT................................................... |
24 |
1 |
6 |
0.67 |
4 |
4 |
0 |
0 |
0 |
| ........................................................................................TTAATCTAAGCTAGGGATGCCGT..................................................... |
23 |
0 |
5 |
0.60 |
3 |
3 |
0 |
0 |
0 |
| ..........................................................................................................................TAGGTTGTAATGGGGCTGCCGTAC.................. |
24 |
0 |
7 |
0.57 |
4 |
4 |
0 |
0 |
0 |
| .....................................................................................................................TGATATAGGTTGTAATGGGACTG........................ |
23 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
| .................................................................................................................TGGCTGATATAGGTTGTAATGGGAC.......................... |
25 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
| ..............................................................................................TAAGCTAGGGATGCCGTATTGGCTGA............................................ |
26 |
0 |
5 |
0.40 |
2 |
2 |
0 |
0 |
0 |
| .............................................................................................CTAAGCTAGGGATGCCGTAT................................................... |
20 |
0 |
5 |
0.40 |
2 |
2 |
0 |
0 |
0 |
| .........................................................................................TAATCTAAGCTAGGGATGCCGTATTGG................................................ |
27 |
0 |
5 |
0.40 |
2 |
2 |
0 |
0 |
0 |
| .........................................................................................TAATCTAAGCTAGGGATGCCGTATTG................................................. |
26 |
0 |
5 |
0.40 |
2 |
2 |
0 |
0 |
0 |
| .........................................................................................TAATCTAAGCTAGGGATGCCGTA.................................................... |
23 |
0 |
5 |
0.40 |
2 |
2 |
0 |
0 |
0 |
| ........................................................................................TTAATCTAAGCTAGGGATGCCGTATA.................................................. |
26 |
1 |
5 |
0.40 |
2 |
2 |
0 |
0 |
0 |
| ..........................................................................................................................TAGGTTGTAATGGGACTGCCGTACTG................ |
26 |
1 |
6 |
0.33 |
2 |
2 |
0 |
0 |
0 |
| ...........................................................................................................................................GCCGTACTGAGTGATAGGG...... |
19 |
0 |
6 |
0.33 |
2 |
0 |
2 |
0 |
0 |
| .................................................................................................................TGGCTGATATAGGTTGTAATGGG............................ |
23 |
0 |
4 |
0.25 |
1 |
0 |
0 |
1 |
0 |
| ......................................................................................TCTTAATCTAAGCTAGGGATGCCG...................................................... |
24 |
0 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
| ..................................................................................................................GGCTGATATAGGTTGTAATGGGAA.......................... |
24 |
2 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
| .....................................................................................TTCTTAATCTAAGCTAGGGATGCCG...................................................... |
25 |
0 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
| .........................................................................................TAATCTAAGCTAGGGATGCCGTATA.................................................. |
25 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ........................................................................................................................................GCTGCCGTACTGAGTGATAGGG...... |
22 |
0 |
5 |
0.20 |
1 |
0 |
1 |
0 |
0 |
| .........................................................................................TAATCTAAGCTAGGGACGCCGTATTGG................................................ |
27 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ..................................................................................................CTAGGGATGCCGTATTGGCTG............................................. |
21 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ................................................................................................................................GTAATGGGGCTGCCGTACTGAGTGA........... |
25 |
0 |
5 |
0.20 |
1 |
0 |
1 |
0 |
0 |
| ....................................................................................................................................TGGGGCTGCCGTACTGAGTGATAGGG...... |
26 |
0 |
5 |
0.20 |
1 |
0 |
1 |
0 |
0 |
| ......................................................................................................................................GGGCTGCCGTACTGAGTGATAGGGG..... |
25 |
0 |
5 |
0.20 |
1 |
0 |
1 |
0 |
0 |
| .........................................................................................TAATCTAAGCTAGGGATGCCGTATT.................................................. |
25 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ..........................................................................................................................AAGGTTGTAATGGGACTGCCGTACTGA............... |
27 |
2 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ....................................................................................................................................TGGGGCTGCCGTACTGAGTGATAGG....... |
25 |
0 |
5 |
0.20 |
1 |
0 |
1 |
0 |
0 |
| ..........................................................................................................................TAGGTTGTAATGGGACTGCCGTACTGA............... |
27 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| .............................................................................................................................GTTGTAATGGGGCTGCCGTACTGAGTGA........... |
28 |
0 |
5 |
0.20 |
1 |
0 |
1 |
0 |
0 |
| ........................................................................................TTAATCTAAGCTAGGGACGCCGTATTG................................................. |
27 |
1 |
5 |
0.20 |
1 |
0 |
1 |
0 |
0 |
| ............................................................................................TCTAAGCTAGGGATGCCGTATTG................................................. |
23 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ..........................................................................................AATCTAAGCTAGGGATGCCGTAT................................................... |
23 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ........................................................................................TTAATCTAGGCTAGGGATGCCGTAT................................................... |
25 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ...................................................................................................TAGGGATGCCGTATTGGCTGAAAAA........................................ |
25 |
2 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| .....................................................................................................................................GGGGCTGCCGTACTGAGTGATAGG....... |
24 |
0 |
5 |
0.20 |
1 |
0 |
1 |
0 |
0 |
| ........................................................................................TTAATCTATGCTAGGGATGCCGT..................................................... |
23 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ...............................................................................................................................................TACTGAGTGATAGGGGTTA.. |
19 |
0 |
6 |
0.17 |
1 |
0 |
1 |
0 |
0 |
| ........................................................................................TTAATCTAAGCTAGGGATGCCG...................................................... |
22 |
0 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
| ..........................................................................................AATCTAAGCTAGGGATGCCG...................................................... |
20 |
0 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
| ........................................................................................TTAATCTAAGCTAGGGATGCCATAT................................................... |
25 |
1 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
| ..........................................................................................AATCTAAGCTAGGGACGCCGTAT................................................... |
23 |
1 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
| ..........................................................................................................................................TGCCGTACTGAGTGATAGGGGT.... |
22 |
0 |
6 |
0.17 |
1 |
0 |
1 |
0 |
0 |
| .........................................................................................TAATCTAAGCTAGGGATGCCG...................................................... |
21 |
0 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
| ...........................................................................................................................AGGTTGTAATGGGGCTGCCGTACTG................ |
25 |
0 |
6 |
0.17 |
1 |
0 |
1 |
0 |
0 |
| ........................................................................................TTAATCTAAGCTAGGGATGCCGC..................................................... |
23 |
1 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
| ..........................................................................................................................................TGCCGTACTGAGTGATAGGG...... |
20 |
0 |
6 |
0.17 |
1 |
0 |
1 |
0 |
0 |
| ..........................................................................................................................TAGGGTGTAATGGGGCTGCCGTACT................. |
25 |
1 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
| ...............................................................................................AAGCTAGGGACGCCGTAT................................................... |
18 |
1 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
| .....................................................................................................................................GGGGCTGCCGTACTGAGCGATAGGG...... |
25 |
1 |
6 |
0.17 |
1 |
0 |
1 |
0 |
0 |
| ............................................................................................TCTAAGCTAGGGATGCCGCATTTG................................................ |
24 |
2 |
6 |
0.17 |
1 |
0 |
0 |
1 |
0 |
| ..........................................................................................................................................TGCCGTACTGAGTGATAGGGGTT... |
23 |
0 |
6 |
0.17 |
1 |
0 |
1 |
0 |
0 |
| ..........................................................................................................................TAGGTTGTAATGGGACTGCCGTACTGAC.............. |
28 |
2 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
| .........................................................................................................................ATAGGTTGTAATGGGACTGCC...................... |
21 |
1 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
| ........................................................................................TTAATCTAAGCTAGGGATGCCGTAA................................................... |
25 |
1 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
| ...........................................................................................................................AGGTTGTAATGGGGCTGCCGTA................... |
22 |
0 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
| ..........................................................................................................................TAGGTTGTGATGGGACTGCCGTAC.................. |
24 |
2 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
| ..........................................................................................................................TAGGTTGTAATGGGGCTGCCGT.................... |
22 |
0 |
7 |
0.14 |
1 |
0 |
1 |
0 |
0 |
| ..........................................................................................................................TAGGTTGTAATGGGGCTGCCG..................... |
21 |
0 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
| .........................................................................................TAATCTAAGCTAGCGATGCCGTAT................................................... |
24 |
1 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
| ........................................................................................TTAATCTAAGCTAGGGATGCC....................................................... |
21 |
0 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
| ..........................................................................................................................TAGGTTGTAATGGGGCTGCCGTA................... |
23 |
0 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
| ............................................................................................................................GGTTGTAATGGGACTGCCGTAC.................. |
22 |
1 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
| ..........................................................................................................................TAGGTTGTAATGGGACTGCCATAC.................. |
24 |
2 |
8 |
0.13 |
1 |
1 |
0 |
0 |
0 |
| ..........................................................................................................................TAGGTTGTAATGGGGCTGCC...................... |
20 |
0 |
8 |
0.13 |
1 |
1 |
0 |
0 |
0 |
| ..........................................................................................................................TAGGTTGTAATGGGGCTGC....................... |
19 |
0 |
8 |
0.13 |
1 |
0 |
1 |
0 |
0 |
| ..........................................................................................................................TAGGTTGTAATGGGACTGC....................... |
19 |
1 |
8 |
0.13 |
1 |
1 |
0 |
0 |
0 |
| .....................................................................................................................................GGTGCTGCCGCACTGAGGGA........... |
20 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |