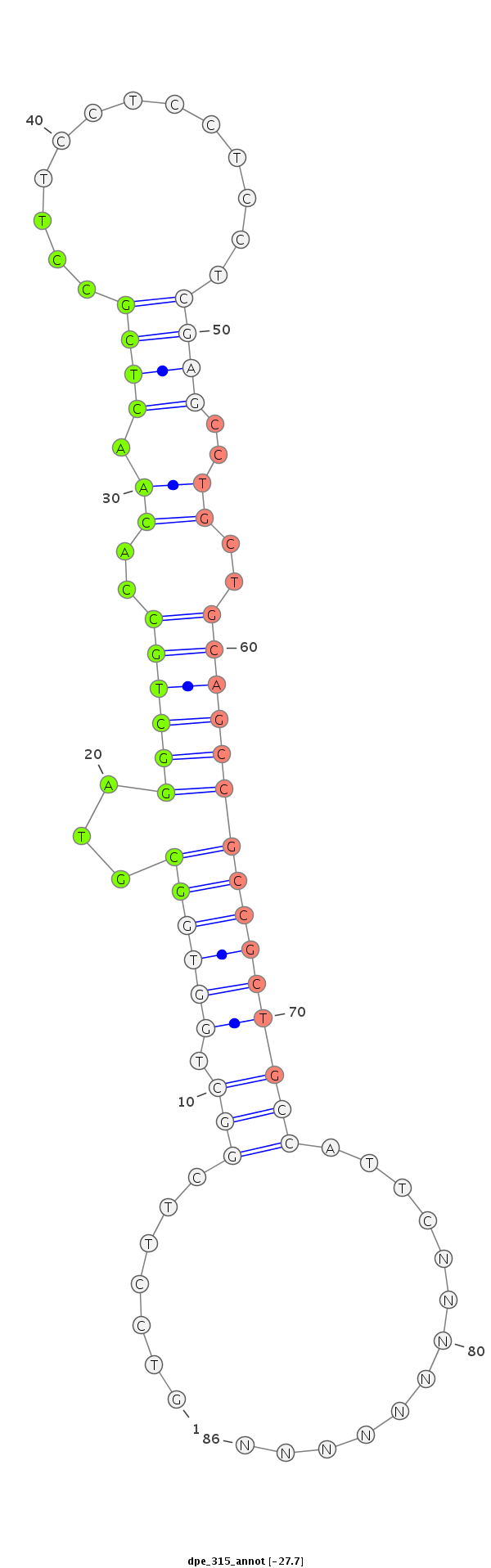
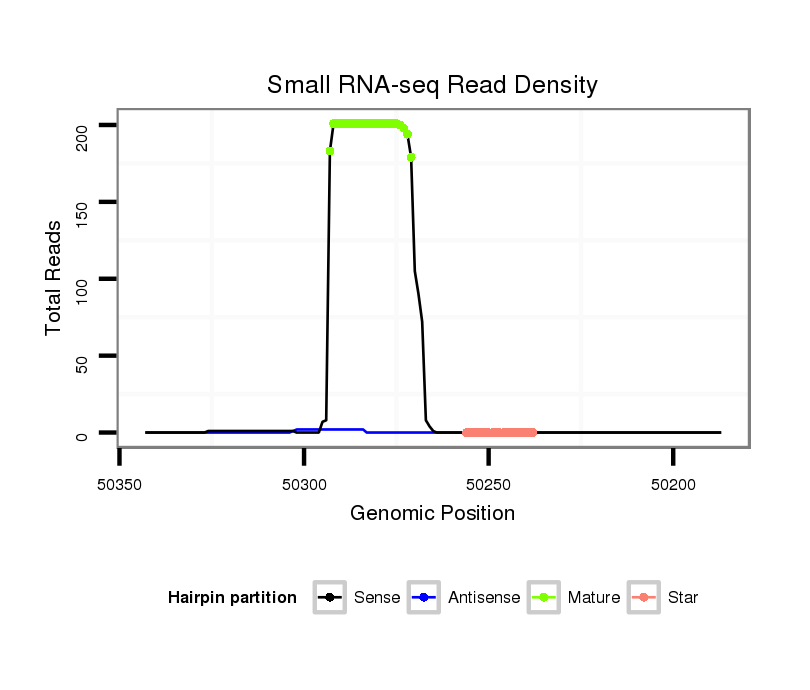
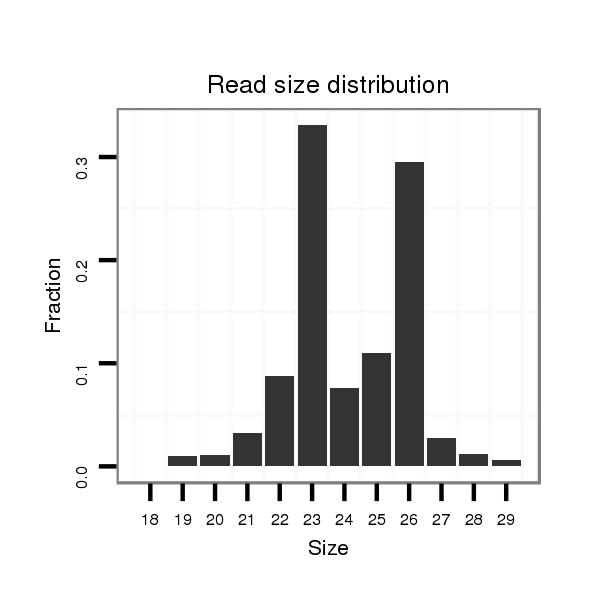
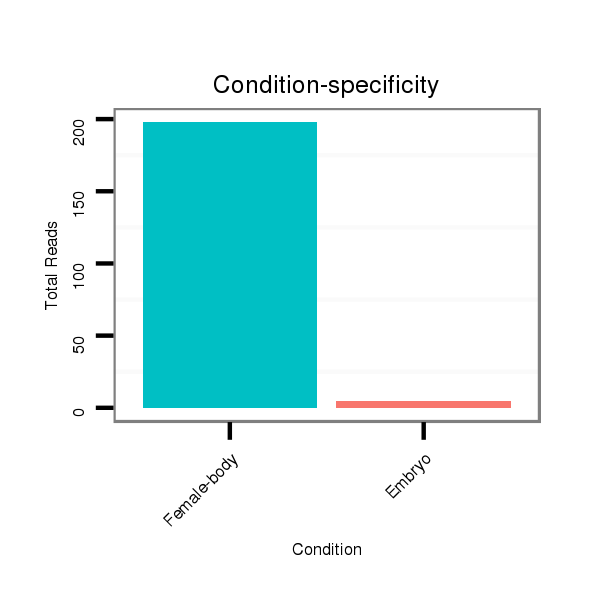
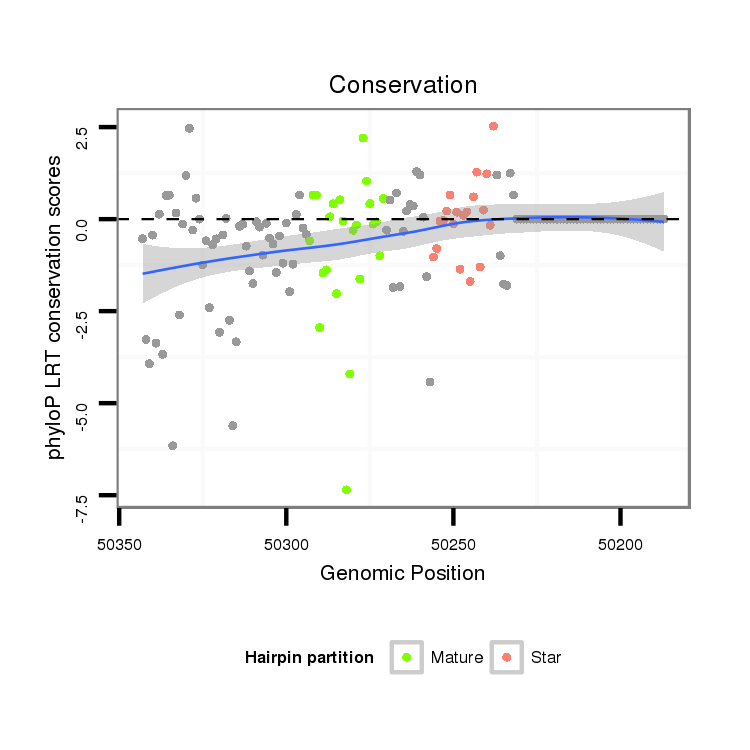
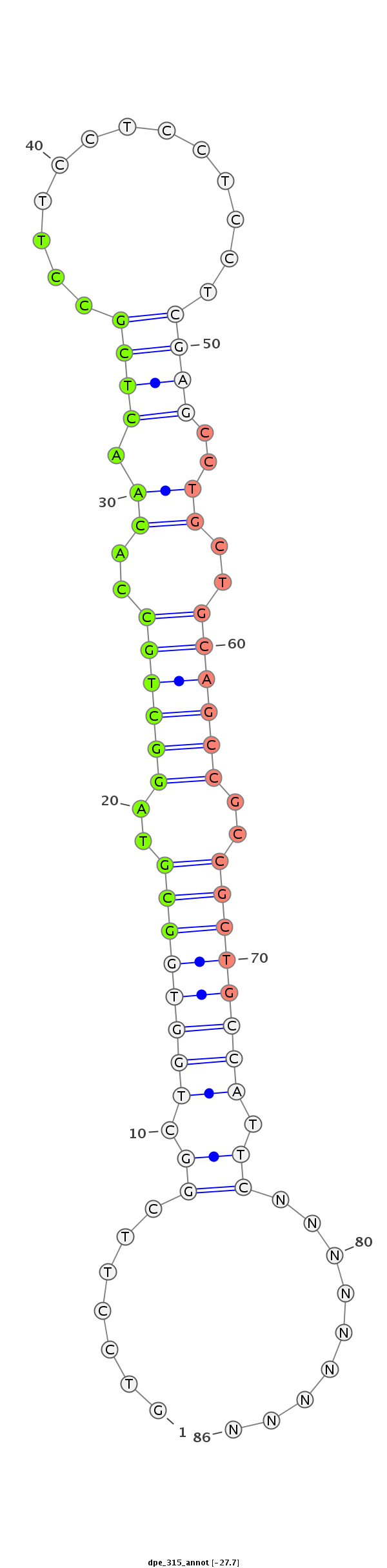
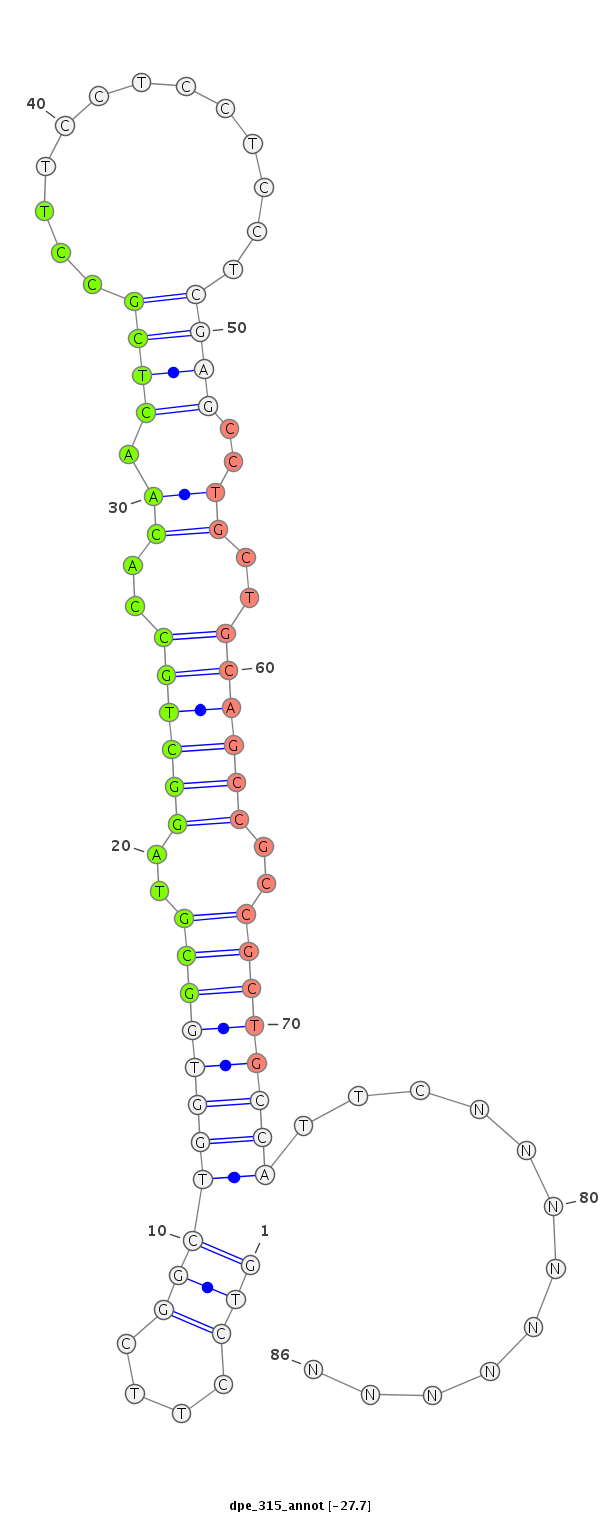
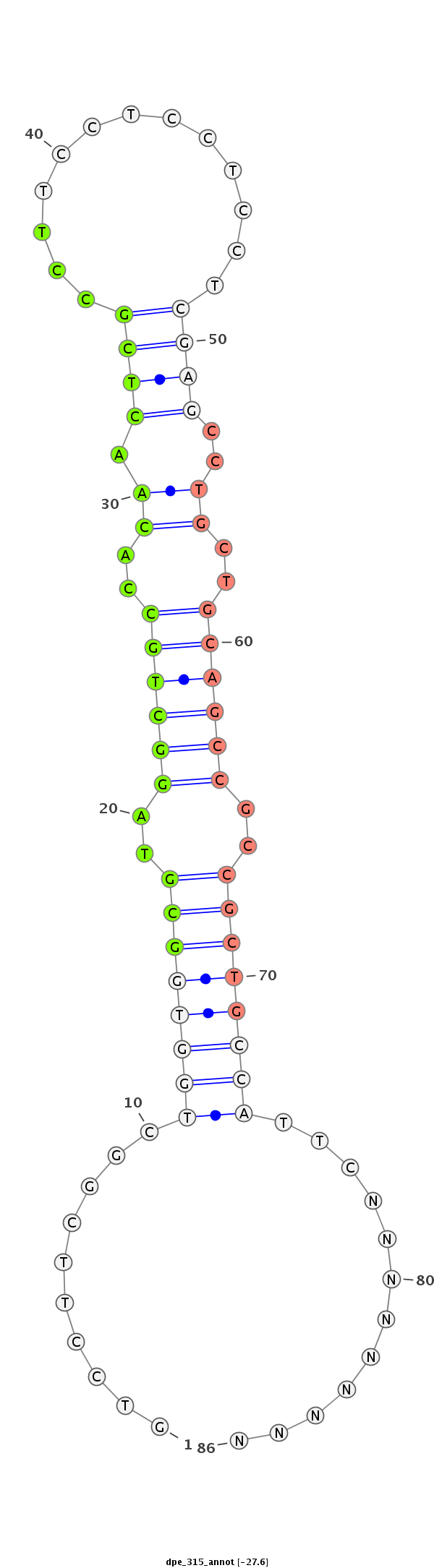
| ..................................................GCGTAGGCTGCCACAACTCGCCT.................................................................................... |
23 |
0 |
4 |
65.00 |
260 |
246 |
14 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTGCCACAACTCGCCTTCC................................................................................. |
26 |
0 |
4 |
59.50 |
238 |
238 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTGCCACAACTCGCCTTC.................................................................................. |
25 |
0 |
4 |
16.75 |
67 |
67 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTGCCACAACTCGCCTT................................................................................... |
24 |
0 |
4 |
13.25 |
53 |
53 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTGCCACAACTCGCC..................................................................................... |
22 |
0 |
4 |
11.00 |
44 |
44 |
0 |
0 |
0 |
0 |
| ...................................................CGTAGGCTGCCACAACTCGCCT.................................................................................... |
22 |
0 |
4 |
6.50 |
26 |
26 |
0 |
0 |
0 |
0 |
| ...................................................CGTAGGCTGCCACAACTCGCCTTCC................................................................................. |
25 |
0 |
4 |
5.50 |
22 |
22 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTGCCACAACTCGCCTTCCT................................................................................ |
27 |
0 |
4 |
4.50 |
18 |
16 |
2 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTGCCACAACTCGC...................................................................................... |
21 |
0 |
4 |
3.50 |
14 |
14 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTGCCACAACTCGCCTTCCTC............................................................................... |
28 |
0 |
4 |
2.50 |
10 |
10 |
0 |
0 |
0 |
0 |
| ................................................TGGCGTAGGCTGCCACAACTCGCCT.................................................................................... |
25 |
0 |
3 |
2.33 |
7 |
6 |
1 |
0 |
0 |
0 |
| ...................................................CGTAGGCTGCCACAACTCGCC..................................................................................... |
21 |
0 |
4 |
2.25 |
9 |
9 |
0 |
0 |
0 |
0 |
| ...................................................CGTAGGCTGCCACAACTCGCCTT................................................................................... |
23 |
0 |
4 |
2.00 |
8 |
8 |
0 |
0 |
0 |
0 |
| ...................................................CGTAGGCTGCCACAACTCGCCTTC.................................................................................. |
24 |
0 |
4 |
2.00 |
8 |
8 |
0 |
0 |
0 |
0 |
| ................................................TGGCGTAGGCTGCCACAACTCGCC..................................................................................... |
24 |
0 |
3 |
2.00 |
6 |
2 |
4 |
0 |
0 |
0 |
| ..................................................GCGTAGGCGGCCACAACTCGCCT.................................................................................... |
23 |
1 |
4 |
1.75 |
7 |
7 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTGCCACAACTCG....................................................................................... |
20 |
0 |
4 |
1.75 |
7 |
6 |
0 |
0 |
1 |
0 |
| ................................................TGGCGTAGGCTGCCACAACTCGC...................................................................................... |
23 |
0 |
3 |
1.33 |
4 |
4 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTGCCACAACTCGCCTTCCTCC.............................................................................. |
29 |
0 |
4 |
1.25 |
5 |
5 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCAGCCACAACTCGCCTTCC................................................................................. |
26 |
1 |
4 |
1.25 |
5 |
5 |
0 |
0 |
0 |
0 |
| ................................................TGGCGTAGGCTGCCACAACTCG....................................................................................... |
22 |
0 |
3 |
1.00 |
3 |
3 |
0 |
0 |
0 |
0 |
| ................................................TGGCGTAGGCTGCCACAACTC........................................................................................ |
21 |
0 |
3 |
1.00 |
3 |
3 |
0 |
0 |
0 |
0 |
| .........................................CGGCTTGTGGCGTAGGCTGCCACAAC.......................................................................................... |
26 |
1 |
2 |
1.00 |
2 |
2 |
0 |
0 |
0 |
0 |
| ...................TCTCGGGGCCCTCGTCGTCCTTT................................................................................................................... |
23 |
1 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
| ..................................................GCGTAGGCGGCCACAACTCGCCTTCC................................................................................. |
26 |
1 |
4 |
1.00 |
4 |
4 |
0 |
0 |
0 |
0 |
| .................CCTCTCGGGGCCCTCGTCGTCCTT.................................................................................................................... |
24 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................................................CGTAGGCTGCCACAACTCGCCTTCCTC............................................................................... |
27 |
0 |
4 |
1.00 |
4 |
4 |
0 |
0 |
0 |
0 |
| .................................................GGCGTAGGCTGCCACAACTCGCCT.................................................................................... |
24 |
0 |
3 |
1.00 |
3 |
3 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCAGCCACAACTCGCCT.................................................................................... |
23 |
1 |
4 |
0.75 |
3 |
3 |
0 |
0 |
0 |
0 |
| ........................................TCGGCTGGTGGCGTAGGCTGCCA.............................................................................................. |
23 |
0 |
4 |
0.75 |
3 |
3 |
0 |
0 |
0 |
0 |
| ........................................TCGGCTGGTGGCGTAGGCTGC................................................................................................ |
21 |
0 |
4 |
0.75 |
3 |
3 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTGCCACAACTC........................................................................................ |
19 |
0 |
4 |
0.75 |
3 |
3 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTGCCACAACTCGCGT.................................................................................... |
23 |
1 |
4 |
0.75 |
3 |
3 |
0 |
0 |
0 |
0 |
| ........................................TCGGCTGGTGGCGTAGGCTGCC............................................................................................... |
22 |
0 |
4 |
0.75 |
3 |
3 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCGGCCACAACTCGCC..................................................................................... |
22 |
1 |
4 |
0.75 |
3 |
3 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCGGCCACAACTCGCCTTC.................................................................................. |
25 |
1 |
4 |
0.75 |
3 |
3 |
0 |
0 |
0 |
0 |
| ................................................TGGCGTAGGCTGCCACAACTCGA...................................................................................... |
23 |
1 |
3 |
0.67 |
2 |
2 |
0 |
0 |
0 |
0 |
| ............TGCCGCCTCTCGGGGCCCTCGTC.......................................................................................................................... |
23 |
0 |
5 |
0.60 |
3 |
3 |
0 |
0 |
0 |
0 |
| ...................................................CGTAGGCTGCCACAACTCGC...................................................................................... |
20 |
0 |
4 |
0.50 |
2 |
2 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTGCCACAACTCGCCTGCC................................................................................. |
26 |
1 |
4 |
0.50 |
2 |
2 |
0 |
0 |
0 |
0 |
| ........................................TCGGCTGGTGGCGTAGGCTGCCACAA........................................................................................... |
26 |
0 |
4 |
0.50 |
2 |
2 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTGCCACAACTCGCCTTG.................................................................................. |
25 |
1 |
4 |
0.50 |
2 |
2 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCCGCCACAACTCGCCTTCC................................................................................. |
26 |
1 |
4 |
0.50 |
2 |
2 |
0 |
0 |
0 |
0 |
| ........................................TCGGCTGGTGGCGTAGGCT.................................................................................................. |
19 |
0 |
4 |
0.50 |
2 |
2 |
0 |
0 |
0 |
0 |
| ........................................TCGGCTGGTGGCGTAGGT................................................................................................... |
18 |
1 |
4 |
0.50 |
2 |
2 |
0 |
0 |
0 |
0 |
| ........................................TCGGCTGGTGGCGTAGGCTG................................................................................................. |
20 |
0 |
4 |
0.50 |
2 |
2 |
0 |
0 |
0 |
0 |
| .....................................................TAGGCTGCCACAACTCGCC..................................................................................... |
19 |
0 |
5 |
0.40 |
2 |
1 |
1 |
0 |
0 |
0 |
| ............TGCCGCCTCTCGGGGCCCTCGTCG......................................................................................................................... |
24 |
0 |
5 |
0.40 |
2 |
2 |
0 |
0 |
0 |
0 |
| ......................................................AGGCTGCCACAACTCGCCT.................................................................................... |
19 |
0 |
5 |
0.40 |
2 |
2 |
0 |
0 |
0 |
0 |
| .................CCTCTCGGGGCCCTCGTCGTCC...................................................................................................................... |
22 |
0 |
6 |
0.33 |
2 |
2 |
0 |
0 |
0 |
0 |
| ................................................TGGCGTAGGCTGCCACAACTA........................................................................................ |
21 |
1 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...............................................GTGGCGTAGGCTGCCACAACTCGCCT.................................................................................... |
26 |
0 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................TCTCGGGACCCTCGTCGTCC...................................................................................................................... |
20 |
1 |
6 |
0.33 |
2 |
0 |
2 |
0 |
0 |
0 |
| .............GCCGCCTCTCGGGGCCCTCGTC.......................................................................................................................... |
22 |
0 |
6 |
0.33 |
2 |
2 |
0 |
0 |
0 |
0 |
| ................................................TGGCGTAGGCTGCCACAACTCGT...................................................................................... |
23 |
1 |
6 |
0.33 |
2 |
2 |
0 |
0 |
0 |
0 |
| ...................TCTCGGGGCCCTCGTCGT........................................................................................................................ |
18 |
0 |
6 |
0.33 |
2 |
2 |
0 |
0 |
0 |
0 |
| .................................................GGCGTAGGCTGCCACAACTCGCCTTCC................................................................................. |
27 |
0 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
| .................................................GGCGTAGGCTGCCACAACTCGCAA.................................................................................... |
24 |
2 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................TCGGCTTGTGGCGTAGGCTGCA............................................................................................... |
22 |
2 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GAGTAGGCTGCCACAACTCGCCT.................................................................................... |
23 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................TCGGCTGGGGGCGTAGGCTGCCACAA........................................................................................... |
26 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTACCACAACTCGCCTTCC................................................................................. |
26 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCCGCCACAACTCGCC..................................................................................... |
22 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................................................CGTAGGCTGCCACAACTCGCCTTGC................................................................................. |
25 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| .................................................CGCGTAGGCTGCCACAACTC........................................................................................ |
20 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTGCCACAACTCGCCA.................................................................................... |
23 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTGCCACAACTCGCCG.................................................................................... |
23 |
1 |
4 |
0.25 |
1 |
0 |
1 |
0 |
0 |
0 |
| ...................................................CGTAGGCTACCACAACTCGCCTTCC................................................................................. |
25 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCGGCCACAACTCG....................................................................................... |
20 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................................................CGTAGGCTCCCACAACTCGCCT.................................................................................... |
22 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| .........................................CGGCTGGTGGCGTAGGCTG................................................................................................. |
19 |
0 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................................................CGTAGGCTGCCACAACTCGCCTTCCT................................................................................ |
26 |
0 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCCGCCACAACTCGCCT.................................................................................... |
23 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...........................................GCTTGTGGCGTAGGCTGCC............................................................................................... |
19 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTGCCACAACTTGCCTTCC................................................................................. |
26 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................TCGGCTTGTGGCGTAGGCTGCCA.............................................................................................. |
23 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................TCGGCTGGTGGCGTAGGA................................................................................................... |
18 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGTACGCTGCCACAACTCGCCT.................................................................................... |
23 |
1 |
4 |
0.25 |
1 |
0 |
1 |
0 |
0 |
0 |
| ..................................................GCTTAGGCTGCCACAACTCGCCTTCC................................................................................. |
26 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................................................CGTAGGCTTCCACAACTCGCCTTCC................................................................................. |
25 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGTTGCCACAACTCGCC..................................................................................... |
22 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................TCGGCTTGTGGCGTAGGCTGC................................................................................................ |
21 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCTTAGGCTGCCACAACTCGCCT.................................................................................... |
23 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGCCTGCCACAACTCGCCTTCC................................................................................. |
26 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................................................CGTAGGCTGCCACAACTCG....................................................................................... |
19 |
0 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGCAGGCTGCCACAACTCGCCT.................................................................................... |
23 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTGCCACAACTCGCAT.................................................................................... |
23 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTGCCACAACTCGCCCTCC................................................................................. |
26 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................TCGTAGGCTGCCACAACTCGCCTTCC................................................................................. |
26 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCGGCCACAACTCGC...................................................................................... |
21 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................TCGGCTTGTGGCGTAGGCTGCCACAA........................................................................................... |
26 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................................................CGTAGGCTGCCACAACTCGCCTTCCA................................................................................ |
26 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCAGCCACAACTCGCCTTC.................................................................................. |
25 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTGCCACAACTCGCCC.................................................................................... |
23 |
1 |
4 |
0.25 |
1 |
0 |
1 |
0 |
0 |
0 |
| ..................................................TCGTAGGCTGCCACAACTCGCCTTC.................................................................................. |
25 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCCTAGGCTGCCACAACTCGCCTTC.................................................................................. |
25 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTTCCACAACTCG....................................................................................... |
20 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................................................CGTAGGCTGCCACAACTCGCCA.................................................................................... |
22 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTGCCACAACTCACCTTCC................................................................................. |
26 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ....................................................GTAGGCTGCCACAACTCGC...................................................................................... |
19 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTGCCACATCTCGCCT.................................................................................... |
23 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| ............TGCCGCCTCTCGGGGCCCTCGT........................................................................................................................... |
22 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................TAGGCTGCCACAACTCGCCTT................................................................................... |
21 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| ............TGCCGCCTCTCGGGGCCCTCGTCC......................................................................................................................... |
24 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................TAGGCTGCGACAACTCGCCTT................................................................................... |
21 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| ............TGCCGCCTATCGGGGCCCTCGTCG......................................................................................................................... |
24 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| ....................................................GTAGGCTGACACAACTCGCCT.................................................................................... |
21 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...........TTGCCGCCTCTCGGGGCCCTCG............................................................................................................................ |
22 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| ............TGCCGCCTCTCGGGGCCCTCGTCGTCC...................................................................................................................... |
27 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...........TTGCCGCCTCTCGGGGCCCTCGTCG......................................................................................................................... |
25 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...........................................................GCCACAACGCGCCTTCCTCCTCCT.......................................................................... |
24 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...........TTGCCGCCGCTCGGGGCCCTCG............................................................................................................................ |
22 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................CAACTCGCCTTCCTCCTCCTCG........................................................................ |
22 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| ....................................................GTAGGCTGCCACAACTCGCCTTCC................................................................................. |
24 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...........TTGCCGCCTCTCGGGGCCCTCGTC.......................................................................................................................... |
24 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGACTGCCACAACTCGCCT.................................................................................... |
23 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................TCGGCTTGTGGCGTAGGCTA................................................................................................. |
20 |
2 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| ............................................................CCACAACTCGCCTTCCTCCTACTC......................................................................... |
24 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| ....................................................GTAGGCTGCCACAACTCGCCT.................................................................................... |
21 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...........TTGCCGCCTCTCGGGGCCCTAG............................................................................................................................ |
22 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| ............TGCGGCCTCTCGGGGCCCTCGTCGTC....................................................................................................................... |
26 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...........................................................GCCACAACGCGCCTTCCTCCTACTC......................................................................... |
25 |
2 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| ............TGCCGCCTCTCGGGGCCCTCGTCGT........................................................................................................................ |
25 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................CTCTCGGGGCCCTCGTCGTCTA..................................................................................................................... |
22 |
2 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
| .............GCCGCCTCTCGGGGCCCTCGTCGTCC...................................................................................................................... |
26 |
0 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
| ....................CTCGGGGCCCTCGTCGTCCT..................................................................................................................... |
20 |
0 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
| ............TGCCGCCTCTCGGGGCCCTC............................................................................................................................. |
20 |
0 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...........................................................GCCACAACTCGCCTTCCTCCT............................................................................. |
21 |
0 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
| ................GCCTCTCGGGGCCCTCGTCGTCC...................................................................................................................... |
23 |
0 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
| .................CCTCTCGGGGCCCTCGTCGTCCT..................................................................................................................... |
23 |
0 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................CAACTCGCCTTCCTCCTC............................................................................ |
18 |
0 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................TCTCGGGGCCCTCGTCGTT....................................................................................................................... |
19 |
1 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..........................................................TGCCACAACTCGCCTTCCTCC.............................................................................. |
21 |
0 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGTAGGCTGCCAAAACTCGTCTTCC................................................................................. |
26 |
2 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................CCTGCTGCAGCCGCCGCTG................................................... |
19 |
0 |
8 |
0.13 |
1 |
0 |
1 |
0 |
0 |
0 |
| ..........................................................TGCCACAAGTCGCCTTCCTCCT............................................................................. |
22 |
1 |
8 |
0.13 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................CCTGCTGCAGCCGCCGCTGT.................................................. |
20 |
1 |
9 |
0.11 |
1 |
0 |
0 |
0 |
1 |
0 |
| .......................................ATCGTCTGGTGGCGTAGGT................................................................................................... |
19 |
3 |
18 |
0.06 |
1 |
0 |
0 |
0 |
0 |
1 |