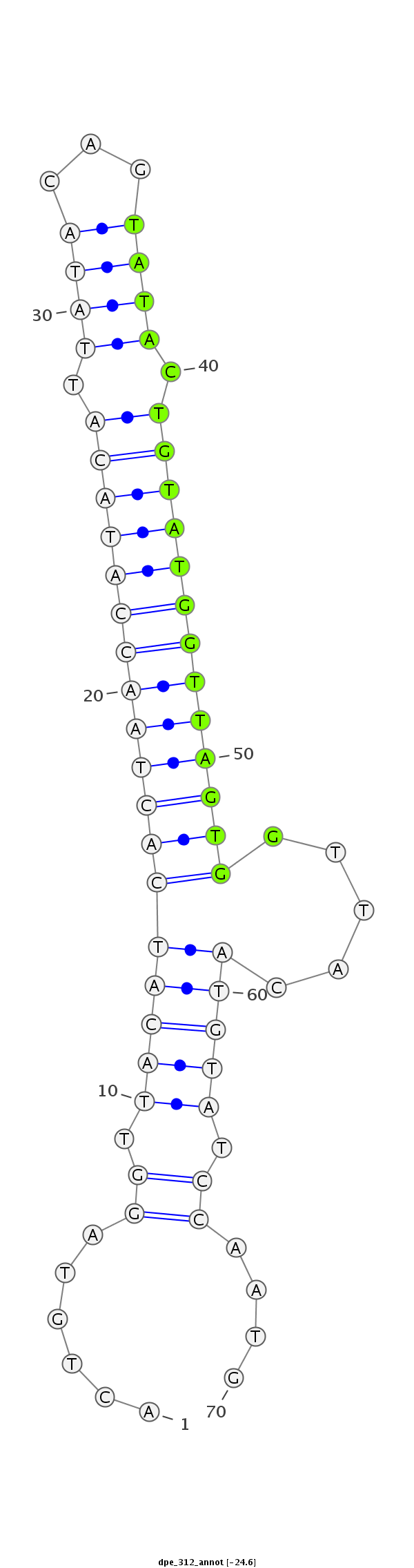
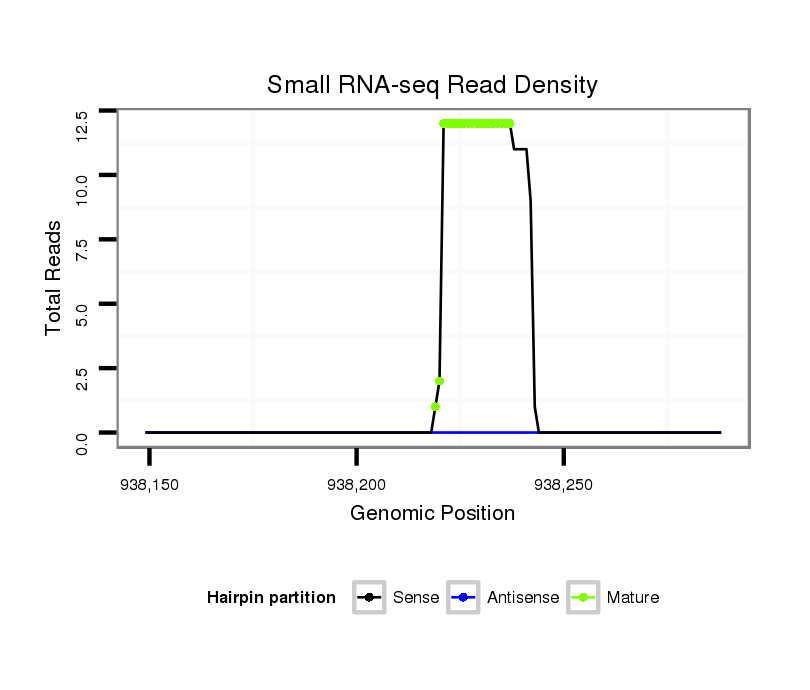
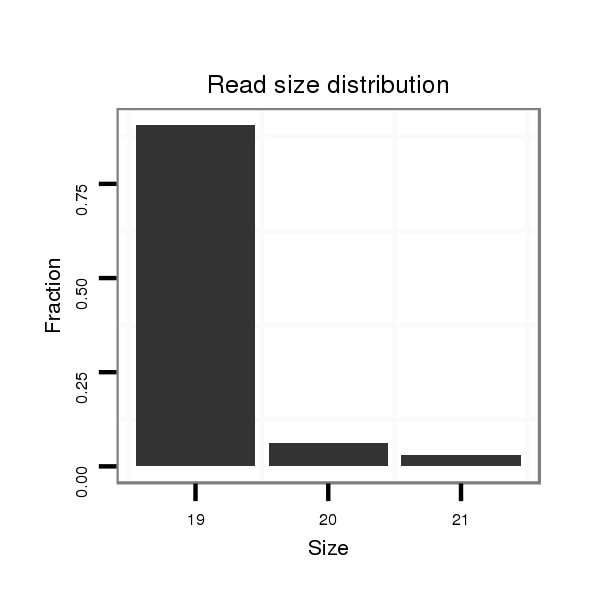
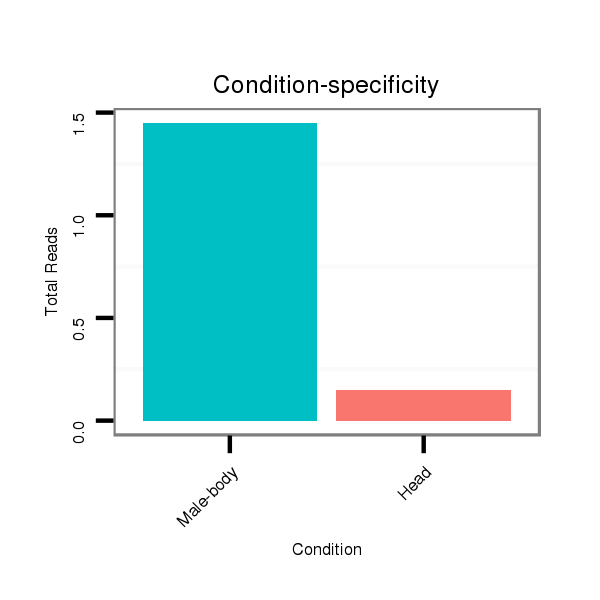
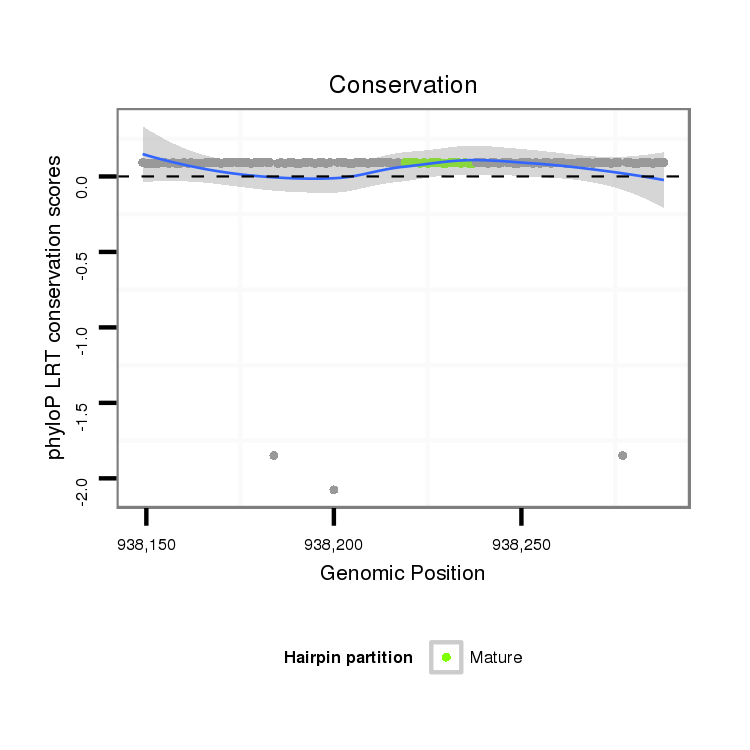
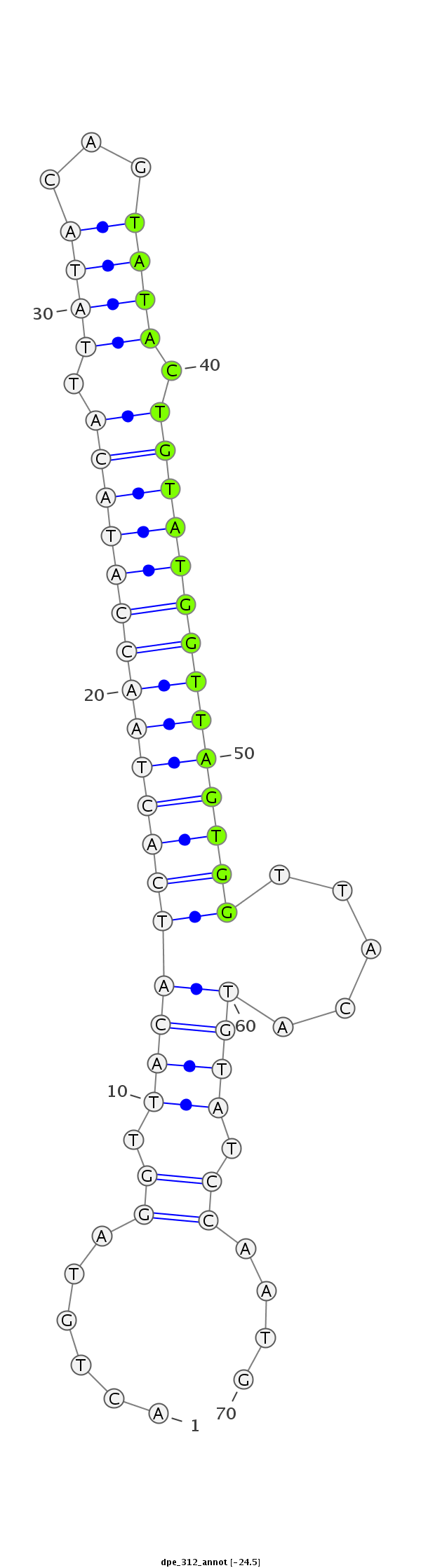
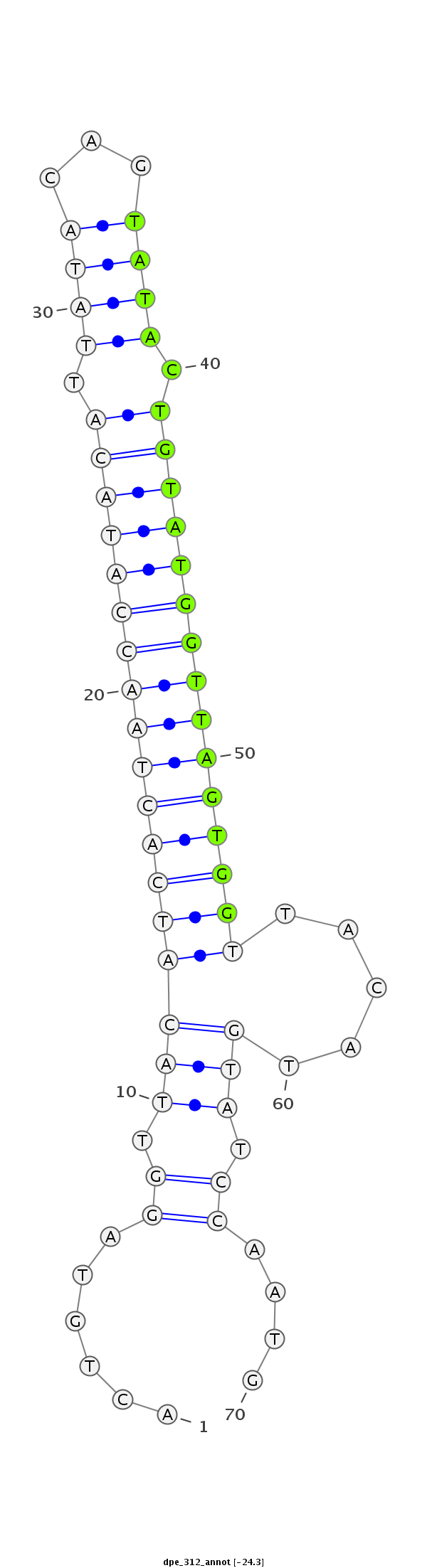
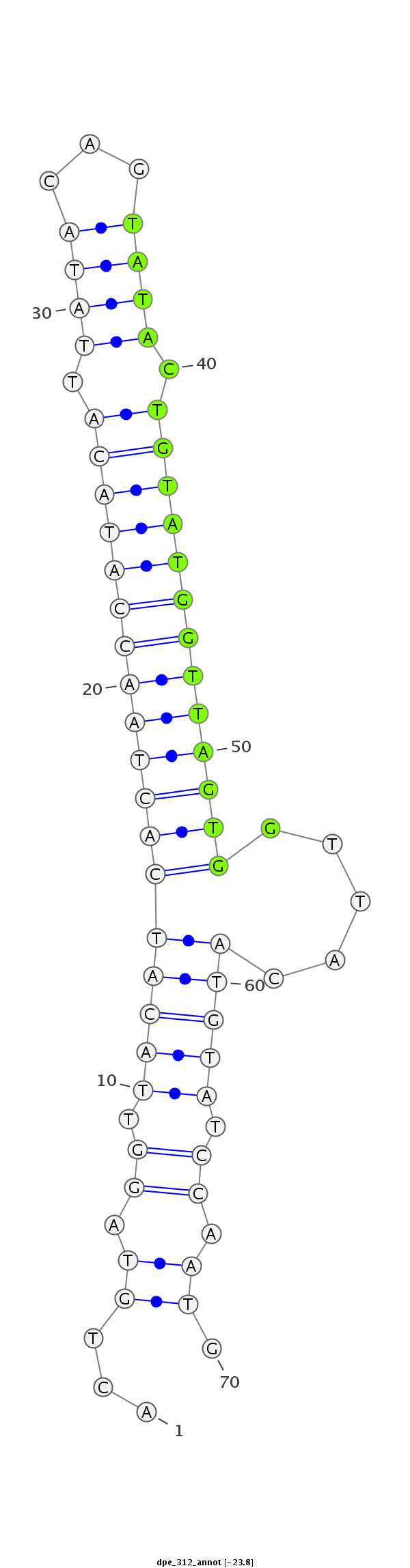
| ........................................................................TACTGTATGGTTAGTGGTTACA.............................................. |
22 |
0 |
2 |
8.50 |
17 |
8 |
3 |
2 |
2 |
2 |
| ........................................................................TACTGTATGGTTAGTGGTTAC............................................... |
21 |
0 |
2 |
1.50 |
3 |
1 |
1 |
1 |
0 |
0 |
| .......................................................................ATACTGTATGGTTAGTGGTTAC............................................... |
22 |
0 |
2 |
1.50 |
3 |
0 |
1 |
0 |
1 |
1 |
| ......................................................................TATACTGTATGGTTAGTGG................................................... |
19 |
0 |
20 |
1.45 |
29 |
29 |
0 |
0 |
0 |
0 |
| ........................................................................TACTGTATGGTTAGTGGTTACAT............................................. |
23 |
0 |
2 |
1.00 |
2 |
2 |
0 |
0 |
0 |
0 |
| ........................................................................TACTGTATGGGTAGTGGGTACA.............................................. |
22 |
2 |
2 |
0.50 |
1 |
0 |
0 |
1 |
0 |
0 |
| ........................................................................TACTGGATGGTTAGTGGTTAC............................................... |
21 |
1 |
2 |
0.50 |
1 |
0 |
0 |
1 |
0 |
0 |
| ......................................................................TATACTGTATGGTTAGTGGTT................................................. |
21 |
0 |
20 |
0.20 |
4 |
4 |
0 |
0 |
0 |
0 |
| .......................................................................ATACTGTATGGTTAGTGGTTA................................................ |
21 |
0 |
18 |
0.17 |
3 |
1 |
1 |
1 |
0 |
0 |
| .......................................................................ATACTGTATGGTTAGTGGTT................................................. |
20 |
0 |
20 |
0.10 |
2 |
0 |
2 |
0 |
0 |
0 |
| ......................................................................TATACTGTATGGTTAGTGGT.................................................. |
20 |
0 |
20 |
0.10 |
2 |
0 |
2 |
0 |
0 |
0 |
| ........................................................................TACTGTATGGTTAGTGGTTA................................................ |
20 |
0 |
18 |
0.06 |
1 |
0 |
0 |
1 |
0 |
0 |
| ......................................................................TATACTGTATGGTTATTGGT.................................................. |
20 |
1 |
20 |
0.05 |
1 |
0 |
0 |
1 |
0 |
0 |
| .....................................................................GTATACTGTATGGTTAGTGGT.................................................. |
21 |
0 |
20 |
0.05 |
1 |
0 |
0 |
1 |
0 |
0 |