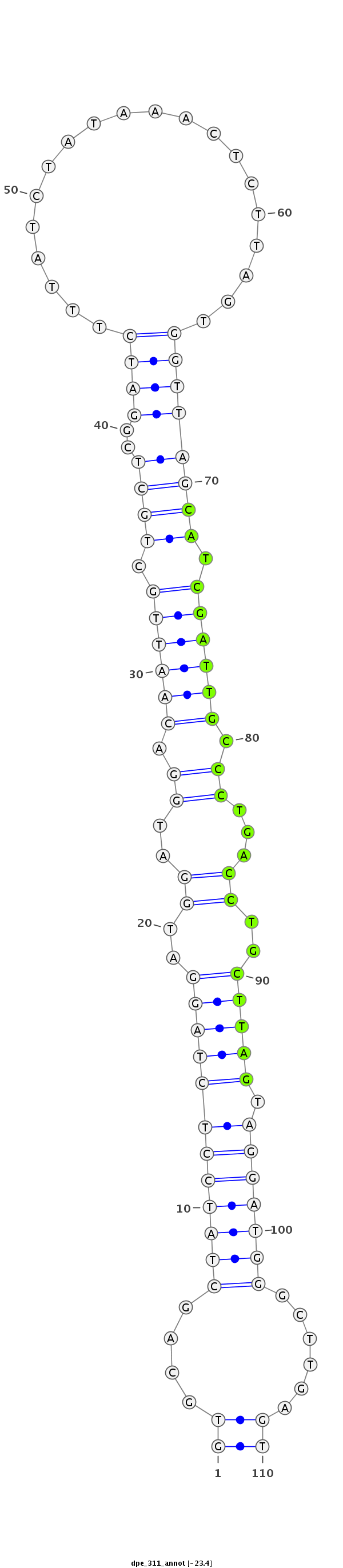

ID:dpe_311 |
Coordinate:scaffold_58:1444-1523 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
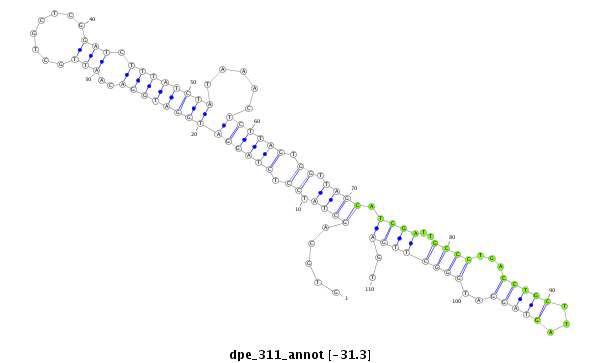
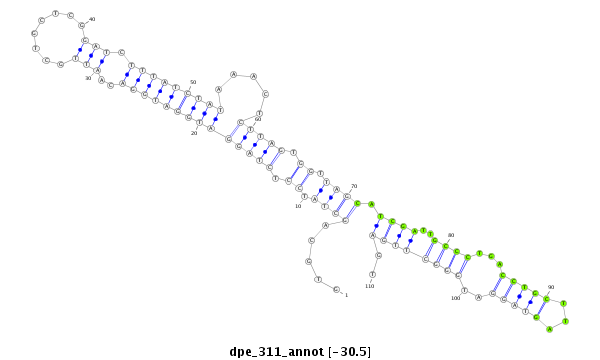
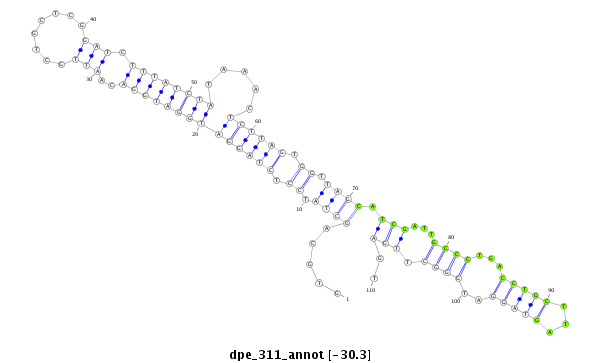
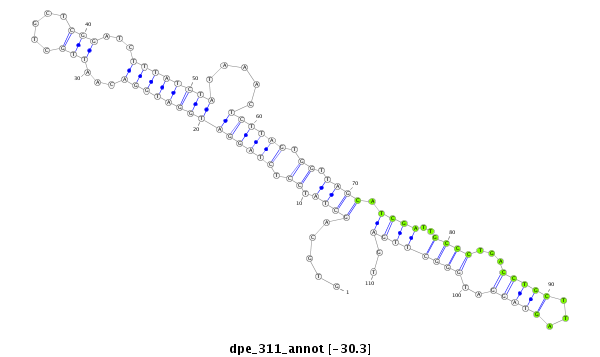
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -31.3 | -30.5 | -30.3 | -30.3 |
 |
 |
 |
 |
intergenic
No Repeatable elements found
| mature | star |
|
ATCCTGGCATCCAGAAAGCTCGCAGATTTCGCCTCGTGCAGCTATCCTCTAGGATGGATGGACAATTGCTGCTCGGATCTTTATCTATAAACTCTTAGTGGTTAGCATCGATTGCCCTGACCTGCTTAGTAGGATGGGCTTGAGTACGTCTGCAAGGAGCTCAAGTACCCATACACCCAC
***********************************((....((((((((((((..((..((.((((((.((((..((((....................)))))))).)))))).))...))..))))).)))))))......))*********************************** |
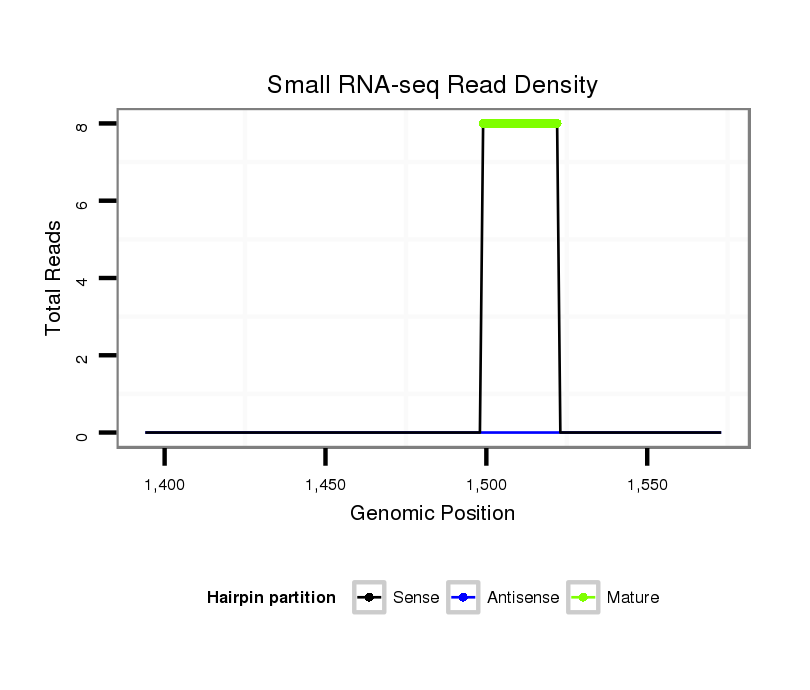
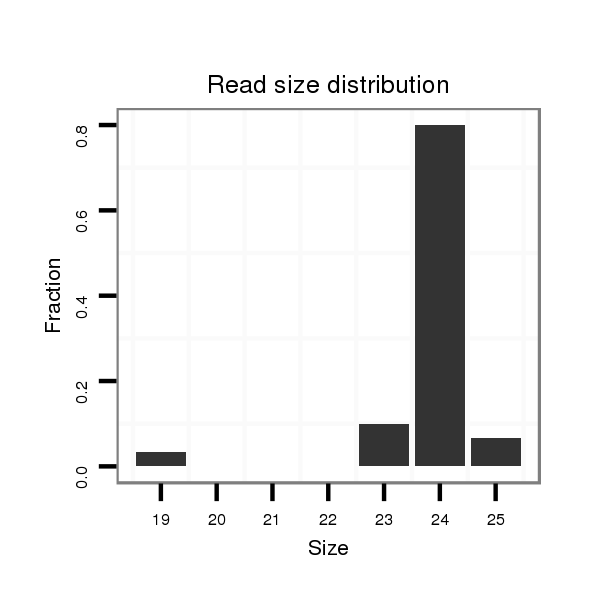
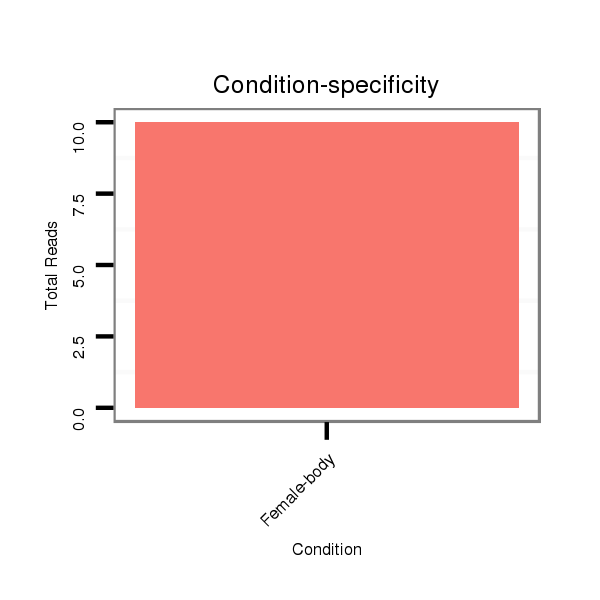
Read size | # Mismatch | Hit Count | Total Norm | Total | M042 female body |
V057 head |
V111 male body |
M021 embryo |
V042 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................CATCGATTGCCCTGACCTGCTTAG................................................... | 24 | 0 | 3 | 8.00 | 24 | 24 | 0 | 0 | 0 | 0 |
| ..................................................................................................TGGTTAGGATCGATTGAC................................................................ | 18 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................TGACATGCATAGTAGGATGGGA......................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................TGGCGTGCTAAGTAGGATGG........................................... | 20 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................CATCGATTGCCCTGACCTGCTTAGT.................................................. | 25 | 0 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..........................................................................................................ATCGATTGCCCTGACCTGCTTAG................................................... | 23 | 0 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 | 0 |
| .........................................................................................................CATCGATTGCCCTGACCTGCTTAGTA................................................. | 26 | 0 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 | 0 |
| .........................................................................................................CATCGATTGCCCTGACCTGCTTA.................................................... | 23 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................CATCGATTGCCCTGACCTG........................................................ | 19 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................CATCGATTGCCCTGACCTGCTTAGTAG................................................ | 27 | 0 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................ATCGATTGCCCTGACCTGCTTAGTAG................................................ | 26 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................CATCGATTTCCCTGACCTGCTTAG................................................... | 24 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................TGCCCTGACCTGCTTAGTAGGAT............................................. | 23 | 0 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................GGTTAAGATCGATTGACCTGA............................................................ | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
|
TAGGACCGTAGGTCTTTCGAGCGTCTAAAGCGGAGCACGTCGATAGGAGATCCTACCTACCTGTTAACGACGAGCCTAGAAATAGATATTTGAGAATCACCAATCGTAGCTAACGGGACTGGACGAATCATCCTACCCGAACTCATGCAGACGTTCCTCGAGTTCATGGGTATGTGGGTG
***********************************((....((((((((((((..((..((.((((((.((((..((((....................)))))))).)))))).))...))..))))).)))))))......))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M042 female body |
V057 head |
|---|---|---|---|---|---|---|---|
| .........................................................................................................GTAGCTAACGGGACTGGACGAAT.................................................... | 23 | 0 | 3 | 0.33 | 1 | 1 | 0 |
| ................................................................................................................ACGGGACTGGACGAATCATC................................................ | 20 | 0 | 9 | 0.11 | 1 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droPer2 | scaffold_58:1394-1573 + | dpe_311 | ATCCTGGCATCCAGAAAGCTCGCAGATTTCGCCTCGTGCAGCTATCCTCTAGGATG-------GATGGACAATT---GCTGCTCGGATCTTTATCTATAA---ACTCTTAGTGGTTAGCATCGATTGCCCTGACCTGCTTAGTAGGATGGGCTTGAGTACGTCTGCAAGGAGCTCAAGTACCCATACA------CCCAC |
| dp5 | XR_group5:404736-404911 - | ATCCTGGCATCCAGAAAGCTCGCAGATTTCGCCTCGTGCAGCTATCCTCTAGGATG-------GATGGACAATT---GCTGCTCGGATCTTTATCTATAA---ACTCTTAGTGGTTAGC----ATTGCCCTGACCTGCTTAGTAGGATGGGCTTGAGTACGTCTGCAAGGAGCTCAAGTACCCATACA------CCCAC | |
| droEle1 | scf7180000490419:22551-22721 + | ATTCTAGCATCTG---------------TCGTCTTGTTCATCCTT---CTAGCCTTGAATACTATAGGAAAATACAAGCTTTACT------TATCTTTGAAGACCTTCTTGCTGGCTGCCTAGATTGCCCTGTGTTGCTTTCCAAGA----CTTTTGTGTGCCATGCAGGTCTTTACGTTCTCACGAACCATTTTTTAT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
Generated: 05/15/2015 at 03:11 PM