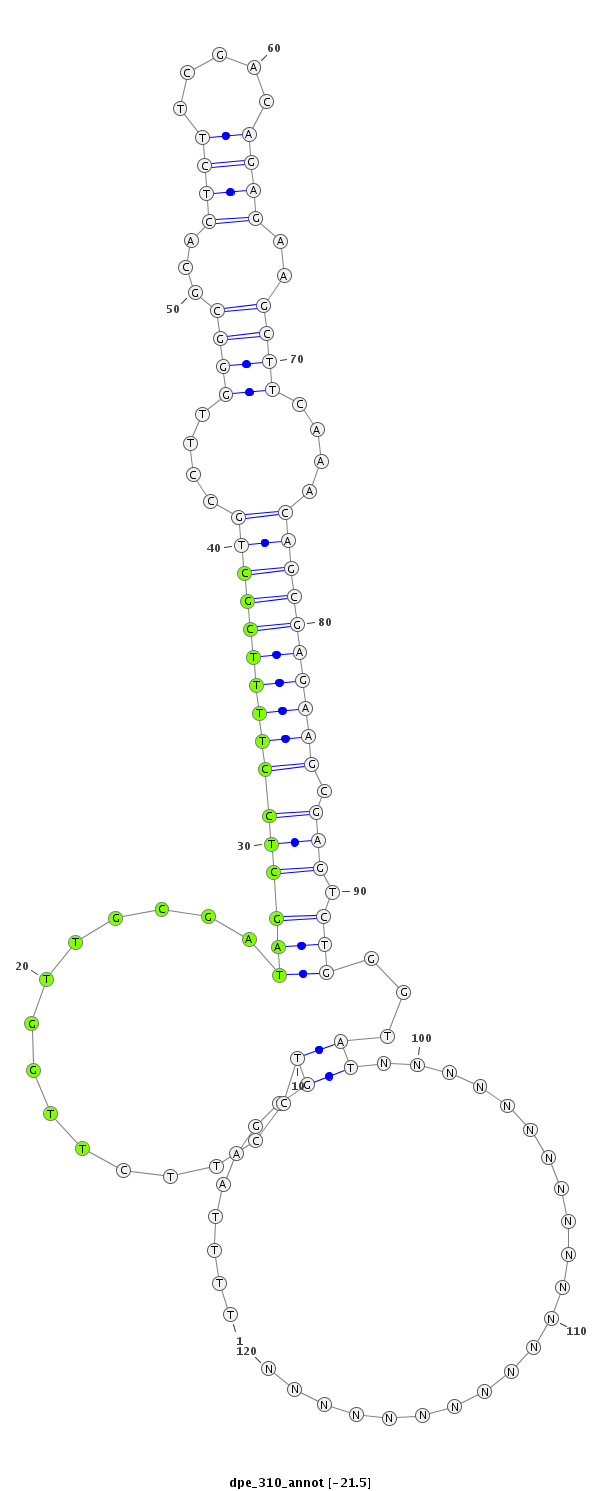
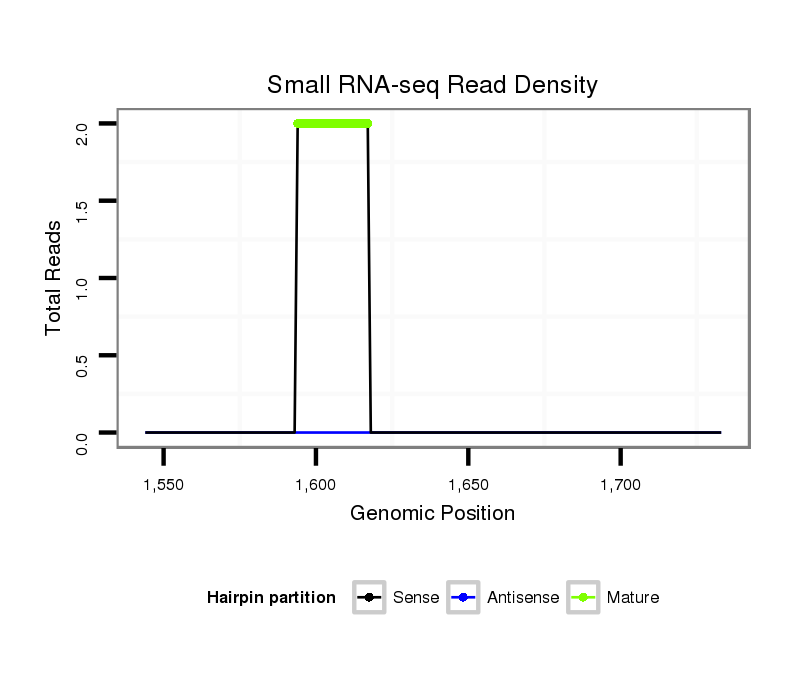
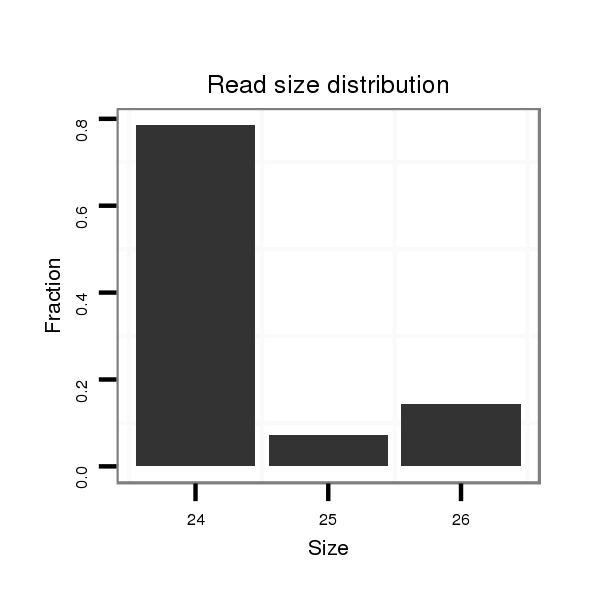
| ..................................................TTGGTTGCGATAGCCCCTTTTCGC.................................................................................................................... |
24 |
1 |
5 |
18.20 |
91 |
89 |
2 |
0 |
0 |
| ..................................................TTGGTTGCGATAGCCCCTTTTCGCTG.................................................................................................................. |
26 |
1 |
5 |
7.40 |
37 |
37 |
0 |
0 |
0 |
| ..................................................TTGGTTGCGATAGCCCCTTTTCG..................................................................................................................... |
23 |
1 |
5 |
4.00 |
20 |
19 |
0 |
1 |
0 |
| ..................................................TTGGTTGCGATAGCCCCTTTTCGCTGC................................................................................................................. |
27 |
1 |
5 |
2.40 |
12 |
12 |
0 |
0 |
0 |
| ..................................................TTGGTTGCGATAGCTCCTTTTCGC.................................................................................................................... |
24 |
0 |
5 |
2.20 |
11 |
11 |
0 |
0 |
0 |
| ..................................................TTGGTTGCGATAGCCCCTTTTC...................................................................................................................... |
22 |
1 |
5 |
1.80 |
9 |
9 |
0 |
0 |
0 |
| ..................................................TTGGTTGCGATAGCCCCTTTTCGCT................................................................................................................... |
25 |
1 |
5 |
1.60 |
8 |
8 |
0 |
0 |
0 |
| ...................................................TGGTTGCGATAGCCCCTTTTCGCTG.................................................................................................................. |
25 |
1 |
6 |
1.00 |
6 |
6 |
0 |
0 |
0 |
| ...................................................TGGTTGCGATAGCCCCTTTTCGC.................................................................................................................... |
23 |
1 |
6 |
1.00 |
6 |
6 |
0 |
0 |
0 |
| ...................................................TGGTTGCGATAGCCCCTTTTCG..................................................................................................................... |
22 |
1 |
6 |
0.83 |
5 |
5 |
0 |
0 |
0 |
| ..................................................TTGGTTGCAATAGCCCCTTTTCGC.................................................................................................................... |
24 |
2 |
5 |
0.60 |
3 |
3 |
0 |
0 |
0 |
| ..................................................TTGGTTGCGATAGCCCCTTTTCGT.................................................................................................................... |
24 |
2 |
5 |
0.40 |
2 |
2 |
0 |
0 |
0 |
| ..................................................TTGGTTGCTATAGCCCCTTTTCGC.................................................................................................................... |
24 |
2 |
5 |
0.40 |
2 |
2 |
0 |
0 |
0 |
| .......................................................TGCGATAGCCCCTTTTCGCTGCCT............................................................................................................... |
24 |
1 |
5 |
0.40 |
2 |
2 |
0 |
0 |
0 |
| ..................................................TTGGTTGCGATAGCTCCTTTTCGCTG.................................................................................................................. |
26 |
0 |
5 |
0.40 |
2 |
2 |
0 |
0 |
0 |
| ......................................................TTGCGATAGCCCCTTTTCGCTGCCT............................................................................................................... |
25 |
1 |
5 |
0.40 |
2 |
2 |
0 |
0 |
0 |
| ............................................................TAGCCCCTTTTCGCTGCCTTGGGCGT........................................................................................................ |
26 |
2 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
| ..................................................TTGGTTGCGATAGCTCCTTTTCGCT................................................................................................................... |
25 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ..................................................TTGGTTGCAATAGCTCCTTTTCGC.................................................................................................................... |
24 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| .........................ACGGGGGCTTTTTTTAGG................................................................................................................................................... |
18 |
2 |
5 |
0.20 |
1 |
0 |
0 |
1 |
0 |
| .......................................................GGCGATAGCCCCTTTTCGCTGCCT............................................................................................................... |
24 |
2 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ..................................................TTGGTTGCGATACCCCCTTTTCG..................................................................................................................... |
23 |
2 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ..................................................TTGGTTGCTATAGCCCCTTTTCGCT................................................................................................................... |
25 |
2 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ..................................................TTGGTTGCGATAGCCCCTTTTCGCAA.................................................................................................................. |
26 |
3 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ..................................................TTGGTTGCGATACCCCCTTTTCGC.................................................................................................................... |
24 |
2 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ..................................................TTGGTTGCGATAGCCCCTTTTCGTTA.................................................................................................................. |
26 |
3 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ..................................................TTGGTTGCGATAGCCCCTTTTCACTGC................................................................................................................. |
27 |
2 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ..................................................TTGGTTGCGATAGCCCCTCTTCGT.................................................................................................................... |
24 |
3 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ..................................................TTGGTTGCGATAGCCCCTTT........................................................................................................................ |
20 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ..................................................TTGGTTGCGATAGCCCCTTTTCGA.................................................................................................................... |
24 |
2 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ................................................TGTTGGTTGCGATAGCCCCTTTT....................................................................................................................... |
23 |
2 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ..................................................TTGGTTGCGATAGCCCCTTTTCGATG.................................................................................................................. |
26 |
2 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ..................................................TTGGTTGCTATAGCCCCTTTTCG..................................................................................................................... |
23 |
2 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| .......................................................TGCGATAGACCCTTTTCGCTGCCT............................................................................................................... |
24 |
2 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ..................................................TTGGTTGCGATAGCCCCTTTTCGCTGCC................................................................................................................ |
28 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ...................................................TGGTTGCGATAGCCCCTTTTCGCTA.................................................................................................................. |
25 |
2 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
| .....................................................GTTGCGATAGCCCCTTTTCG..................................................................................................................... |
20 |
1 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
| ...................................................TGGTTGCGATAGCCCCTTTTC...................................................................................................................... |
21 |
1 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
| ...................................................TGGTTGCGGTAGCCCCTTTTCGCTG.................................................................................................................. |
25 |
2 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
| ...................................................TGGTTGCGATAGCCCCTTTTCGA.................................................................................................................... |
23 |
2 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
| ...................................................TGGTTGCGATAGCCCCTTTTCGCTGC................................................................................................................. |
26 |
1 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
| .....................................................TTTGCGATAGCCCCTTTTCGCTG.................................................................................................................. |
23 |
2 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
| ....................................................GGTTGCGATAGCCCCTTTTCGC.................................................................................................................... |
22 |
1 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
| ...................................................TGGTTGCGCTAGCCCCTTTTCGCTG.................................................................................................................. |
25 |
2 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
| ...................................................TGGTTGCGATACCCCCTTTTCGCTGC................................................................................................................. |
26 |
2 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
| ..............................................CTGCTTAGTTGCGATAAC.............................................................................................................................. |
18 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
1 |