
ID:dpe_299 |
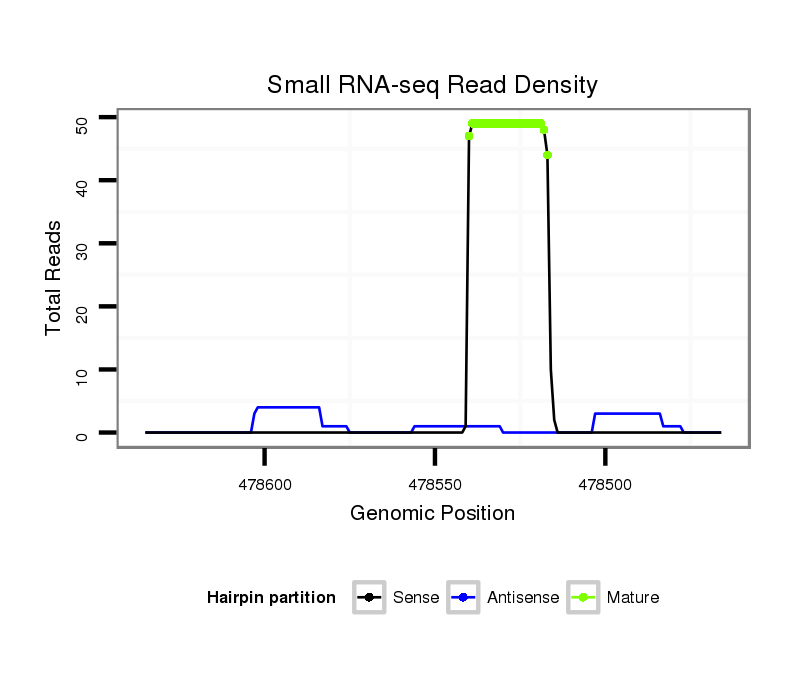
Coordinate:scaffold_3:478516-478585 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -23.8 | -23.5 | -23.3 |
 |
 |
 |
intergenic
No Repeatable elements found
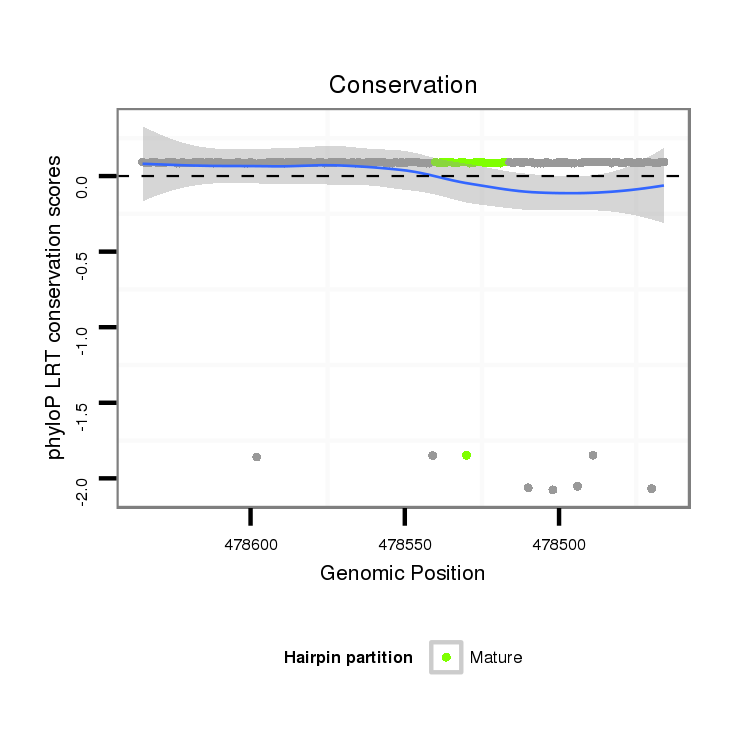
| mature | star |
|
ACCTAGGCTATCGTCGGATAGTACTTCCGGGACCCTGTCGGACTAAGAATTAATACGAAGGTCTTTGCTCTGCTAGATACCGACAGTTATCGCCAAGAGTACCTAGGCTATCGCCGGATAGTACTGACGGGACCCTGTCGGACTAAGAATTACTACGAAGGTCTTTGCTA
***********************************((((((((((....((....(((.(((((.((((((....((((........))))....))))))...))))).)))...)))))).))))))......*********************************** |
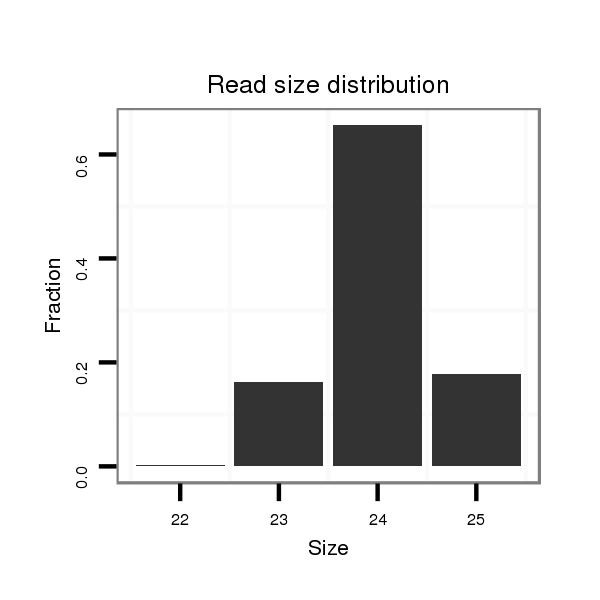
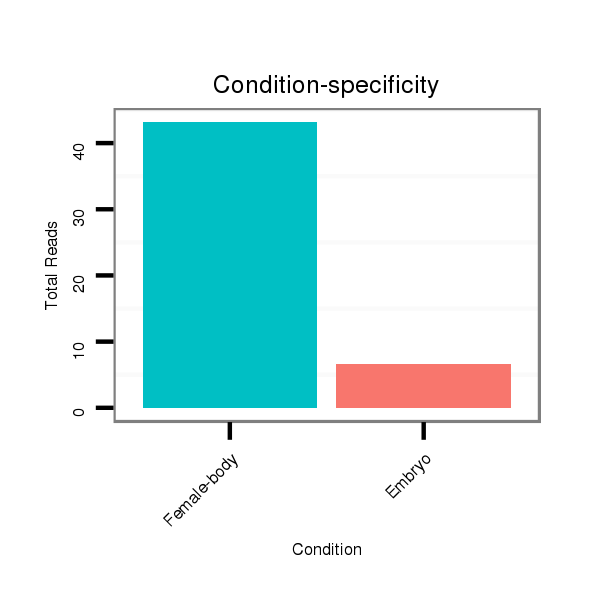
Read size | # Mismatch | Hit Count | Total Norm | Total | M042 female body |
M021 embryo |
V042 embryo |
|---|---|---|---|---|---|---|---|---|
| ...............................................................................................AGAGTACCTAGGCTATCGCCGGAT................................................... | 24 | 0 | 5 | 32.00 | 160 | 135 | 11 | 14 |
| ...............................................................................................AGAGTACCTAGGCTATCGCCGGATA.................................................. | 25 | 0 | 5 | 8.80 | 44 | 36 | 3 | 5 |
| ...............................................................................................AGAGTACCTAGGCTATCGCCGGA.................................................... | 23 | 0 | 20 | 4.95 | 99 | 99 | 0 | 0 |
| ...............................................................................................AGAGTACCTAGGCTATCGCCGGATAG................................................. | 26 | 0 | 5 | 2.80 | 14 | 7 | 6 | 1 |
| ................................................................................................GAGTACCTAGGCTATCGCCGGAT................................................... | 23 | 0 | 6 | 2.00 | 12 | 12 | 0 | 0 |
| ...............................................................................................AGAGTACCTAGGCTATCGCCGGATAA................................................. | 26 | 1 | 8 | 1.13 | 9 | 6 | 3 | 0 |
| ..............................................................................................AAGAGTACCTAGGCTATCGCCGG..................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...............................................................................................AGAGTACCGAGGCTATCGCCGGAT................................................... | 24 | 1 | 14 | 0.93 | 13 | 13 | 0 | 0 |
| ...............................................................................................AGAGTACCAAGGCTATCGCCGGAT................................................... | 24 | 1 | 5 | 0.80 | 4 | 4 | 0 | 0 |
| ................................................................................................GAGTACCTAGGCTATCGCCGGATA.................................................. | 24 | 0 | 6 | 0.67 | 4 | 4 | 0 | 0 |
| ................................................................................................GAGTACCTAGGCTATCGCCGGATAG................................................. | 25 | 0 | 6 | 0.67 | 4 | 4 | 0 | 0 |
| ...............................................................................................AGAGTACCAAGGCTATCGCCGGATA.................................................. | 25 | 1 | 5 | 0.20 | 1 | 1 | 0 | 0 |
| ...............................................................................................AGAGTACCTAGGCTATCGCCGGCT................................................... | 24 | 1 | 5 | 0.20 | 1 | 0 | 0 | 1 |
| ...............................................................................................AGAGTACCTAGGCTATCGCCGGATT.................................................. | 25 | 1 | 10 | 0.20 | 2 | 2 | 0 | 0 |
| ...............................................................................................AGAGTACCTAGGATATCGCCGGATA.................................................. | 25 | 1 | 5 | 0.20 | 1 | 0 | 1 | 0 |
| ...............................................................................................AGAGTACCTAGCCTATCGCCGGATA.................................................. | 25 | 1 | 6 | 0.17 | 1 | 1 | 0 | 0 |
| ..................................................................................................GTACCTAGGCTATCGCCGGATAG................................................. | 23 | 0 | 6 | 0.17 | 1 | 1 | 0 | 0 |
| ...............................................................................................AGAGTACCTAGGCTATCGCCGGGT................................................... | 24 | 1 | 6 | 0.17 | 1 | 0 | 0 | 1 |
| .................................................................................................AGTACCTAGGCTATCGCCGGATA.................................................. | 23 | 0 | 6 | 0.17 | 1 | 1 | 0 | 0 |
| ...............................................................................................AGAGTACCGAGGCTATCGCCGGATA.................................................. | 25 | 1 | 12 | 0.17 | 2 | 2 | 0 | 0 |
| .................................................................................................AGTACCTAGGCTATCGCCGGAT................................................... | 22 | 0 | 6 | 0.17 | 1 | 1 | 0 | 0 |
| ........................................................................................ATCGACAAGACAACCTAGG............................................................... | 19 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 |
| ...............................................................................................AGAGTACCTAGGCTATCGCCGGCTAA................................................. | 26 | 2 | 8 | 0.13 | 1 | 0 | 1 | 0 |
| .............................GGACCCTGTCGCACCACGA.......................................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 2 | 0 | 0 |
| ...............................................................................................AGAGTACCAAGGCTATCGCCGGATT.................................................. | 25 | 2 | 10 | 0.10 | 1 | 1 | 0 | 0 |
| ...............................................................................................GGAGTACCTAGGCTATCGCCGGAT................................................... | 24 | 1 | 10 | 0.10 | 1 | 1 | 0 | 0 |
| ...............................................................................................AGAGTACCGAGGCTATCGCCGGATAG................................................. | 26 | 1 | 12 | 0.08 | 1 | 1 | 0 | 0 |
| ...............................................................................................AGAGTACCTATGCTATCGCCGGAT................................................... | 24 | 1 | 16 | 0.06 | 1 | 1 | 0 | 0 |
| ...............................................................................................AGAGTACCAAGGCTATCGCCGGA.................................................... | 23 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ...............................................................................................AGAGTACCCAGGCTATCGGCGGAT................................................... | 24 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ..............................................................................................AGCAGTACCTAGGCTACCGCCGGA.................................................... | 24 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 |
|
TGGATCCGATAGCAGCCTATCATGAAGGCCCTGGGACAGCCTGATTCTTAATTATGCTTCCAGAAACGAGACGATCTATGGCTGTCAATAGCGGTTCTCATGGATCCGATAGCGGCCTATCATGACTGCCCTGGGACAGCCTGATTCTTAATGATGCTTCCAGAAACGAT
***********************************((((((((((....((....(((.(((((.((((((....((((........))))....))))))...))))).)))...)))))).))))))......*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M042 female body |
M021 embryo |
V042 embryo |
V111 male body |
V050 head |
V057 head |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................GGGACAGCCTGATTCTTAAT...................................................................................................................... | 20 | 0 | 20 | 2.80 | 56 | 48 | 3 | 0 | 5 | 0 | 0 |
| ....................................................................................................................................GGGACAGCCTGATTCTTAAT.................. | 20 | 0 | 20 | 2.80 | 56 | 48 | 3 | 0 | 5 | 0 | 0 |
| ....................................................................................................................................GGGACAGCCTGATTCTTAATGATGCT............ | 26 | 0 | 20 | 1.15 | 23 | 17 | 5 | 1 | 0 | 0 | 0 |
| .................................GGACAGCCTGATTCTTAAT...................................................................................................................... | 19 | 0 | 20 | 1.05 | 21 | 14 | 5 | 2 | 0 | 0 | 0 |
| ...............................................................................GGCTGTCAATAGCGGTTCTCATGGAT................................................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................GGGACAGCCTGATTCTTAATTATGCTTC.............................................................................................................. | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................GGACAGCCTGATTCTTAATGATGCT............ | 25 | 0 | 20 | 0.65 | 13 | 13 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................ACAGCCTGATTCTTAATGATGCT............ | 23 | 0 | 20 | 0.45 | 9 | 9 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................GGACAGCCTGATTCTTAAT.................. | 19 | 0 | 20 | 0.35 | 7 | 0 | 5 | 2 | 0 | 0 | 0 |
| ...............................TGGGACAGCCTGATTCTTAA....................................................................................................................... | 20 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................CCCTGGGACAGCCTGATTCTTAAT.................. | 24 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................TGGGACAGCCTGATTCTTAA................... | 20 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................CCCTGGGACAGCCTGATTCTTAAT...................................................................................................................... | 24 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................TGGTACAGCCTGATTCTTAAT.................. | 21 | 1 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................TGGTACAGCCTGATTCTTAAT...................................................................................................................... | 21 | 1 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................GGGACAGCCTGATTCTTAA................... | 19 | 0 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| ................................GGGACAGCCTGATTCTTAA....................................................................................................................... | 19 | 0 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................TTGCGGATCCGATAGCGGCCTATCAT............................................... | 26 | 3 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................GGACAGCCTGATTCTTAA................... | 18 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................GGGACAGCCTGATTCTTAATGATGC............. | 25 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................ACAGCCTGATTCTTAATGATGC............. | 22 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .................................GGACAGCCTGATTCTTAA....................................................................................................................... | 18 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ................................GGGACAGCCTGCTTCTTAAT...................................................................................................................... | 20 | 1 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................CGGATCCGATAGCGGCCTATCATGA............................................. | 25 | 1 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................AGGCCCCGGGAGAGCTTGAT............................................................................................................................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................GGACAGCCTGATTCTTAATGATGC............. | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................CCTGCGACAGCCTGATTCTTAAT...................................................................................................................... | 23 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................GCGGATCCGATAGCGGCCTATCAT............................................... | 24 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................GGGACAGCCTGATTCTGAAT.................. | 20 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................GACAGCCTGATTCTTAATGATGCT............ | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................GGGACAGCCTGGTTCTTAAT.................. | 20 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................TGGACAGCCTGATTCTTAAT...................................................................................................................... | 20 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................GGACAGCCTGATTCTTAATGATGCA............ | 25 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................AGACGAACTTTGGCTATCA................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................................................CCTGCGACAGCCTGATTCTTAAT.................. | 23 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droPer2 | scaffold_3:478466-478635 - | dpe_299 | ACCTAGGCTATCGTCGGATAGTACTTCCGGGACCCTGTCGGACTAAGAATTAATACGAAGGTCTTTGCTCTGCTAGATACCGACAGTTATCGCCAAGAGTACCTAGGCTATCGCCGGATAGTACTGACGGGACCCTGTCGGACTAAGAATTACTACGAAGGTCTTTGCTA------ |
| dp5 | 2:18140966-18141141 - | ACCTAGGCTATCGTCGGATAGTACTTCCGGGACCCTGCCGGACTAAGAATTAATACGAAGGTCTTTGCTCTGCTAGATACCGACAGTTATCGCCGAGAGTACCTAAGCTATCGCCGGATAGTACTTACGGGACACTGTCGGCCTAAAAATTACTACGAAGGTCTTGGCTATGCTGA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
Generated: 05/15/2015 at 03:09 PM