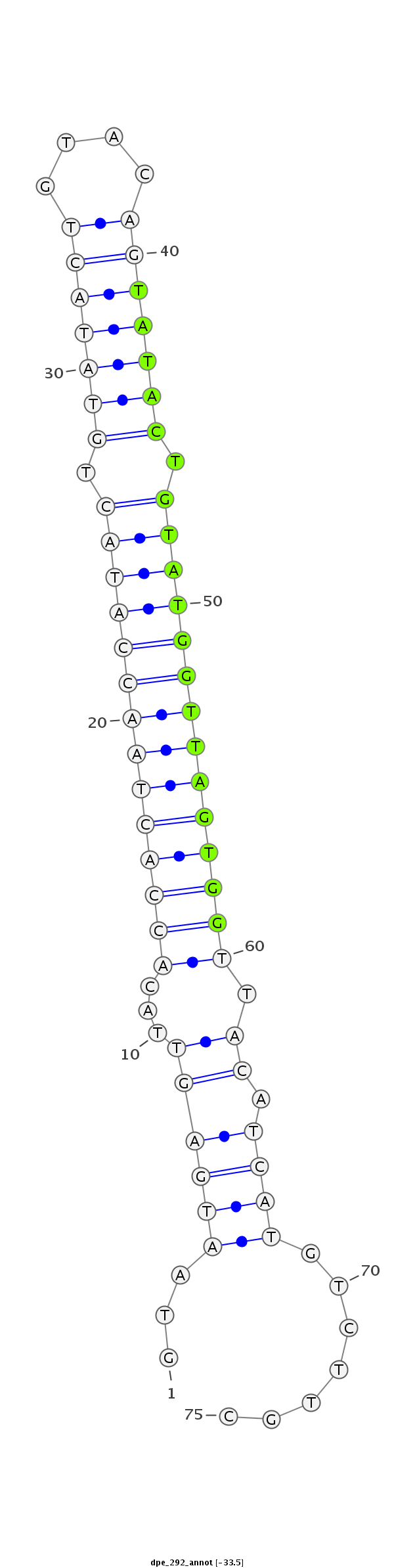



ID:dpe_292 |
Coordinate:scaffold_15:330109-330153 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -33.3 | -33.2 |
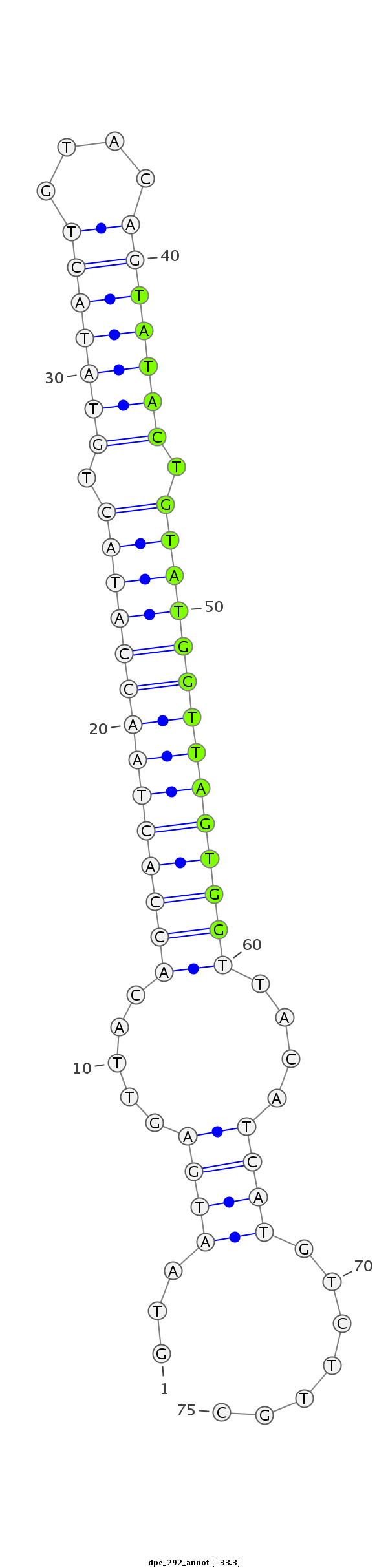 |
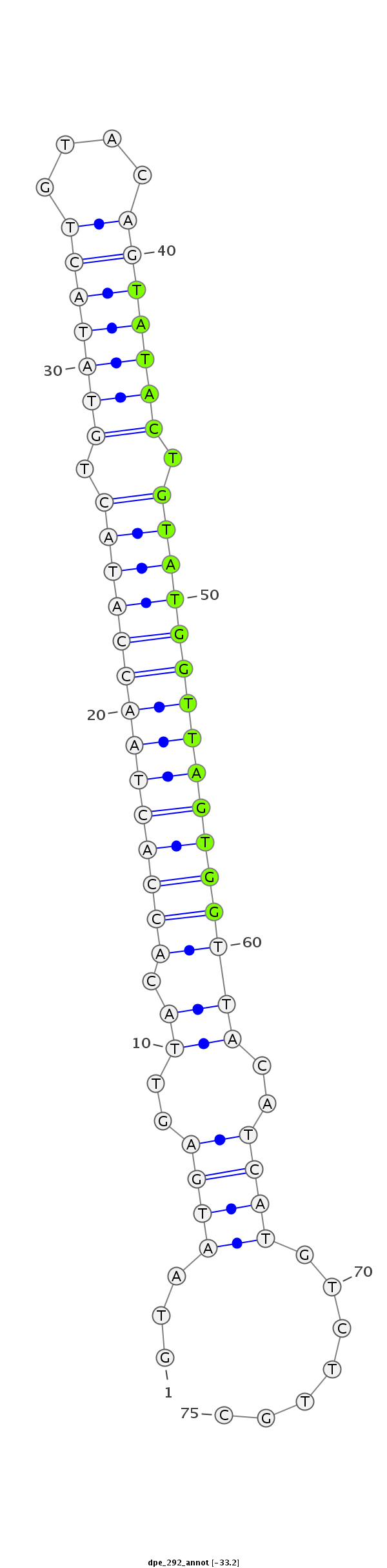 |
intergenic
No Repeatable elements found
|
ACACTGATCCCGAACCGAAACACGTGCGTGTAACAGTAATGAGTTACACCACTAACCATACTGTATACTGTACAGTATACTGTATGGTTAGTGGTTACATCATGTCTTGCTCACACACCTTTCATATTTGAATACTTTTTATAGT
***********************************...((((((...((((((((((((((.(((((((....))))))).)))))))))))))).)).)))).......*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V111 male body |
V050 head |
V057 head |
M042 female body |
V042 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................TACTGTATGGTTAGTGGTTACA.............................................. | 22 | 0 | 2 | 8.50 | 17 | 8 | 3 | 2 | 2 | 2 |
| .............................................................................TACTGTATGGTTAGTGGTTAC............................................... | 21 | 0 | 2 | 1.50 | 3 | 1 | 1 | 1 | 0 | 0 |
| ............................................................................ATACTGTATGGTTAGTGGTTAC............................................... | 22 | 0 | 2 | 1.50 | 3 | 0 | 1 | 0 | 1 | 1 |
| ...........................................................................TATACTGTATGGTTAGTGG................................................... | 19 | 0 | 20 | 1.45 | 29 | 29 | 0 | 0 | 0 | 0 |
| .............................................................................TACTGTATGGTTAGTGGTTACAT............................................. | 23 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .............................................................................TACTGTATGGGTAGTGGGTACA.............................................. | 22 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................TACTGGATGGTTAGTGGTTAC............................................... | 21 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................TATACTGTATGGTTAGTGGTT................................................. | 21 | 0 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 |
| ............................................................................ATACTGTATGGTTAGTGGTTA................................................ | 21 | 0 | 18 | 0.17 | 3 | 1 | 1 | 1 | 0 | 0 |
| ............................................................................ATACTGTATGGTTAGTGGTT................................................. | 20 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ...........................................................................TATACTGTATGGTTAGTGGT.................................................. | 20 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| .............................................................................TACTGTATGGTTAGTGGTTA................................................ | 20 | 0 | 18 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................TATACTGTATGGTTATTGGT.................................................. | 20 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................GTATACTGTATGGTTAGTGGT.................................................. | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
|
TGTGACTAGGGCTTGGCTTTGTGCACGCACATTGTCATTACTCAATGTGGTGATTGGTATGACATATGACATGTCATATGACATACCAATCACCAATGTAGTACAGAACGAGTGTGTGGAAAGTATAAACTTATGAAAAATATCA
***********************************...((((((...((((((((((((((.(((((((....))))))).)))))))))))))).)).)))).......*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V057 head |
V111 male body |
|---|---|---|---|---|---|---|---|
| .............................................TGTGGTGATTGGTATGACAT................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ............................................ATGTGGTGATTGGTATGACAT................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droPer2 | scaffold_15:330059-330203 + | dpe_292 | ACACTGATCCCGAACCGAAACACGTGCGTGTAACAGTAATGAGTTACACCACTAACCATACTGTATACTGTACAGTATACTGTATGGTTAGTGGTTACATCATGTCTTGCTCACACACCTTTCATATTTGAATACTTTTTATAGT |
| dp5 | XL_group1a:7973125-7973218 + | ACACTGATCCGGAACCGAAACACGTGCGTGTAACAGTAATGAGTTGCACC---------------------------------------------------ATGTCTTGGTCACACACCCTTCATATTTGAATACTTTTTATGGT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
Generated: 05/15/2015 at 03:08 PM