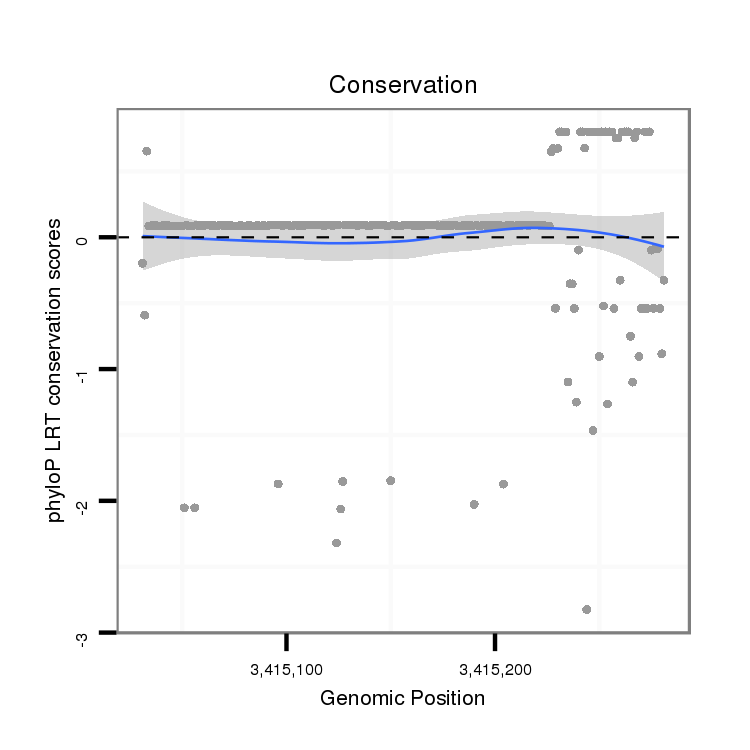
ID:dpe_2106 |
Coordinate:scaffold_8:3415081-3415231 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dper\GL16384-cds]; exon [dper_GLEANR_1805:1]; intron [Dper\GL16384-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- AGCGCATCGGTGACCGCGGGAACGCAAGTGTTACTCCTAGCAACCCGAGGGTGCGACCTGCTAAATAGAAAGGAAGAGTTTTATAAAGTAACACTCCTTTAATGATATGCAAATATTCGGAAGGCTATATCTCAGATAAATAGGACCAGATCGGTGAAGGACCAACTCTCCAGACTGTCGACTATGAAAATGAGGCCAATTTTTGGCATATTTAATGATGATTTTTTGCGAAAATCGTGAAAAAATGGATG **************************************************...(((..(((.......(((((.((.(((.........))).)))))))........))).....)))...(((.....((((.....((((.....((.((((...(((.......)))..))))))..)))).....)))))))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V111 male body |
V057 head |
M021 embryo |
|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................................TTTGGGCATATTTCATGTTGAT............................. | 22 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 |
| ..................................................................................................................................................................................................................................TGCGAAAATCGTGAAAAAATGGATG | 25 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 |
| ................................................................................................................................................ACCAGGTCGGTGAAGGAC......................................................................................... | 18 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ..................................................................................................................................................................................................................................TGCGAAAATCGTGAAAAAATGG... | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
|
TCGCGTAGCCACTGGCGCCCTTGCGTTCACAATGAGGATCGTTGGGCTCCCACGCTGGACGATTTATCTTTCCTTCTCAAAATATTTCATTGTGAGGAAATTACTATACGTTTATAAGCCTTCCGATATAGAGTCTATTTATCCTGGTCTAGCCACTTCCTGGTTGAGAGGTCTGACAGCTGATACTTTTACTCCGGTTAAAAACCGTATAAATTACTACTAAAAAACGCTTTTAGCACTTTTTTACCTAC
**************************************************...(((..(((.......(((((.((.(((.........))).)))))))........))).....)))...(((.....((((.....((((.....((.((((...(((.......)))..))))))..)))).....)))))))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M042 female body |
V057 head |
|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................ATATCCTGGTCTAGCCAGTTCCTGGT....................................................................................... | 26 | 2 | 3 | 0.33 | 1 | 1 | 0 |
| ................................................................................................................................................................TGGTCGAGTGGTCTGACGGC....................................................................... | 20 | 3 | 8 | 0.13 | 1 | 0 | 1 |
| ........................................................................................................................TTCCGTTGTAGAGGCTATTT............................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 |
| ...................................................................................................................................................................................................................................CGCTTTTAGCACTTTTTTACCTA. | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droPer2 | scaffold_8:3415031-3415281 + | dpe_2106 | AGCGCATCGGTGACCGCGGGAACGCAAGTGTTACTCCTAGCAACCCGAGGGTGCGACCTGCTAAATAGAAAGGAAGAGTTTTATAAAGTAACACTCCTTTAATGATATGCAAATATTCGGAAGGCTATATCTCAGATAAATAGGACCAGATCGGTGAAGGACCAACTCTCCAGACTGTCG------------------ACTATGAAAATGAGGCCAATTTTTGGCATATTT-----------AATGATGATTTTTTGCGAAAATC-GTGAAAAAATGGATG |
| dp5 | 4_group2:471252-471531 - | dps_1449 | AGCGCATCGGTGACCGCGGGCACGCCAGTGTTACTCCTAGCAACCCGAGGGTGCGACCTGCTAAAAAGAAAGGAAGAGTTTTATAAAGTAACAGTATTTTAATGATATGCAAATATTCGAAAGGCTATATCTCAGATAAATAGGACCAGATCGGTGAAGTACCAACTCTCCAGTCTGTCGAACGGCATCAAAATGTCAACTATGAAAATGAGGCCAATTTTTGGCAAATTTAATGATGTAACCATCATGATTATTTGCGAAAATC-GTGAAAAAATGGATG |
| droBia1 | scf7180000301760:24777-24839 - | CCC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A-TTTTCGCAATTTTTGTGA---GTGCATCATGAATTTTTGCAAAAATG-GTGAAAAATTAAAAA | |
| droTak1 | scf7180000414562:99286-99342 - | C---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CATTTTTTAATTATTT-----------GGATCATCATTTTTAGCGAAAAGTGGTCTTATATAAATGG |
| Species | Read alignment | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
|||||||||||||||||||||||||||||||||
| dp5 |
|
|||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||
| droTak1 |
|
Generated: 06/08/2015 at 05:11 PM