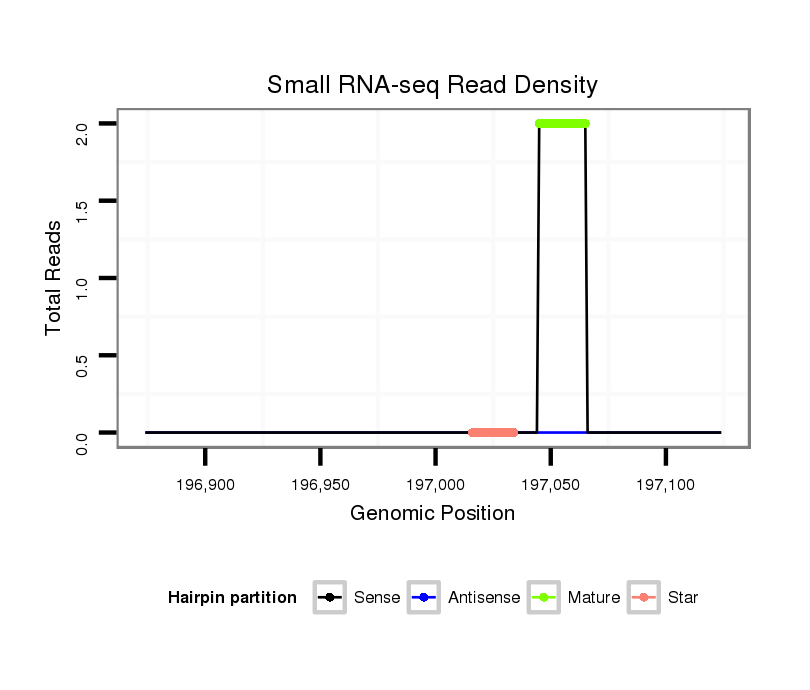
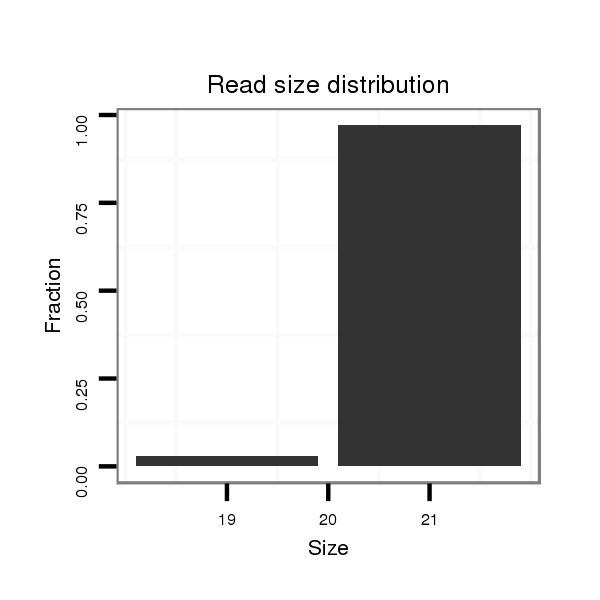
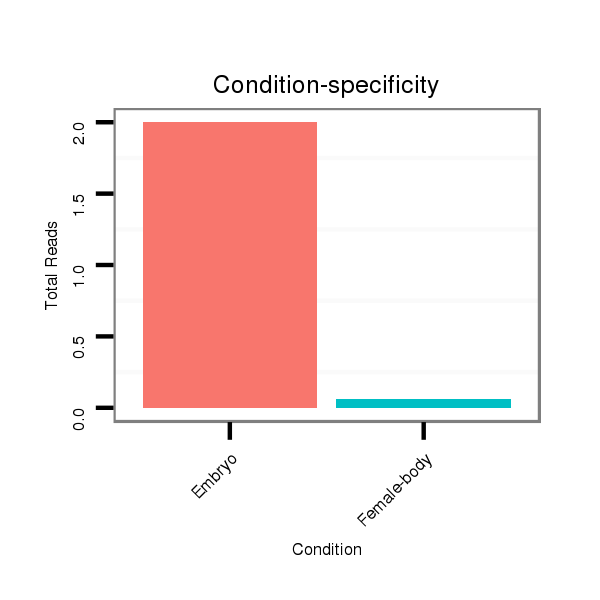
| droPer2 |
scaffold_61:196874-197124 + |
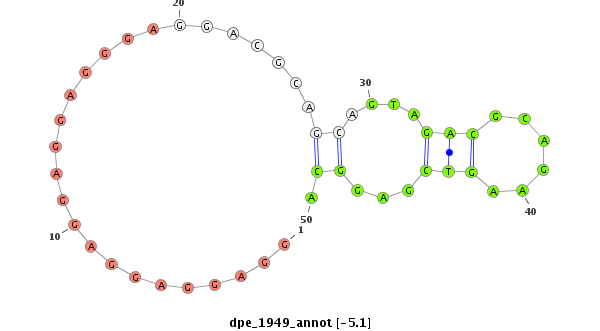
dpe_1949 |
AGCAGCAGCAGCAGAAGCAGA--A------------------------------------------GCAG---------CAGTCAC---C---------------CACCA--------------------TCGATGATAA-------------------GGGATTG-----------------CAGCACTCGGAAGAT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------ACAAAGGACACAGATA--------------------------------------CAGATACGCAGAGCATTCGAGACA-CAG------------CAGA----------------------------------------------------------------------ATCTGAGCTT----------------GAGGCTG-AAGCTGAGGAGGAGGAGGA--------------------------------------------------------GG-AG--------GGAGGACGCAGCAGTAGACGC---------AGAAGT------------CGAGGCAG-TCATCATGGGTAAAGAT-AGACGAGATCCCGTTTTGCATA-CCGTTCGTCGAGTGCCGTC |
| dp5 |
XR_group6:9916862-9917095 + |
|
AGCAGAAGCAGCAGTCAC-----------------------------------------------------------------------C---------------CACCA--------------------TCGATGATAA-------------------GGGATTG-----------------CAGCACTCGGAAGAT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------ACAAAGGACACAGATA--------------------------------------CAGATACGCAGAGCATTCGAGACA-CAG------------CAGA----------------------------------------------------------------------ATCTGAGCTT----------------GAGGCAG-AAGCTGAGGAGGAGGAG----------------------------------------------------------GAGGA--------GGAGGACGCAGCAGTAGACGC---------AGAAGT------------CGAGGCAG-TCATCATGG-TAAAGAT-AGACGAGACCCCGTTTTACGTA-CCGTTCGTCGAGTGCCGTT |
| droWil2 |
scf2_1100000004762:1177067-1177337 + |
|
AACAGCTTCAGCTTCAGCATC--A------------------------------------------GCTT---------CAGCTTC---A---------------C---GACA--------------TAATCGATGATAA-------------------CGACG--AG--------------------CTC-----------------------TTAGAC---AAAGACATACAGGAATCACAAGTGCAGCAGCAGCAGC---------------AGCAGCAGCA------------------GACC-ACA--A----------------AGAGCGACAGTAGATCCAGGACTAGAACCAGAAATACAAACGAAACGGAAT--------------------------------------CAGTTACAAGAAACGAACTATTTA-CA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAGAGGAGCA--AGAAGACGCAGAAGTTGACGC---------GGAAGT------------CGAGGCTG-TCATTATGG-TAAAAATCAAAAGAAA---------------------------------- |
| droVir3 |
scaffold_13049:20458661-20458847 + |
|
AACAACAGCAGCAGCAGCGGC--A------------------------------------------GCAG---------CAGCAGC---A---------------GGACG--------------------ACGACAATAA-------------------TATTG--CA--------------------GAT-----------------------CTAGAC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCTGAGGAGGA---------GGAGCAGCAGCAACAGCAGGCGGGGGAC-----------------------------------------------------------------------GACGGGGAAGAGGAAGAGGAC--------GAGGAAGAAGAGGACGA--------CGAGGAGGAGCAGGACGAGGTCGAGGAAGAAGAAGC------------GGAGGAGG---------T--------------------------------------------------- |
| droMoj3 |
scaffold_6680:6627234-6627430 - |
|
-GCAGCAACAGCTCAAGAATT--T------------------------------------------G------------------------C---ACAAAGAACAC---TTTG--------------ACATCGATGACAACGCACTAGCAGCGACAGAGACAGAGG-----------------C--------------------------------A---------------------------GAGCAGCAGCAGCAGC---------------AGCAGCAGCA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAGCAACAGCAACAGCAGAGAGATAAT-----------------------------------------------------------------------TC-----------------------GAGCAGAGCCACA---GCAGA--------GGAGGACGCAGCAGTTGACGC---------GGAAGT------------CGAGGCAG-TCATCATGG--------------------------------------------------- |
| droGri2 |
scaffold_15074:5980298-5980500 - |
|
AGCAGCAGCAGCAGCAGCAGAGCA------------------------------------------GCAG---------CAGA----TCGCGA---CAACGATCAGAGG-----------------------------------------------------------------------------------------CACTGAATACAGAGCCACAGACACA----------------------------------------------GCCA-T---------------------------AGCCATAGC-------------------CAC----AGCAAGAGTCAGA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCC-AGGGCCAAGA----------CAA-CGACAACGACAGC-----AGCA-----------------------------------------------------GCGGC--------GGCGGCATCAGCAGCAGCAGC---------AGCAGC------------AGCAGCAG-CTACAGCAA-TGAAAAT-TGCTGAGA---------------------------------- |
| droAna3 |
scaffold_13266:10910354-10910594 + |
|
AGCATCAGCAGCAGAGGCAGA--G------------------------------------------GCAG---------CCACCGA-----------------------------------------------------------------------------------AGCGGCGTT-------------------------------------------------------------------------------G--ATGGCCGAAGC----GGTGGCTGTA------------------GCT-TACC-------------------TGA----GCGACGAGGAA----------------------------------------------------------------------------------------------G-AGGAAGAGCCCGAGACTGGGGGAGAGCCTGATCCGGACGTTGAGGAAGATGCAGTGGA-------------------------------ATTCGCTGTC----------------GAAGAC------------------------------------GCTGGCGAGGATCAGGATGAA--------GGGGCTCCAGAGGACGA--------GGAGGAAGCCGCCGGCGAAGC---------TGGCGA------------GGAAGACG-CTG-TAAGT--------------------------------------------------- |
| droBip1 |
scf7180000396384:24026-24191 - |
|
AGCAGCAACATCAGCAGCAGC--A------------------------------------------GCAA---------CAACACT---C---------------G--------------------------------------------------------------GAGCAACTGCAGCA--------------------------GGAG--------------------------------------------TA--ACT------------------------AGCGATACT-GCAGCAGGTCTAGC-------------------GGG----CGGAGGAGGGA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGGGAA----------------GAGGCAG-AGACAGGGGAGTAGGAGGA-----------G--------------------------------------------GTGGT--------GGAGGAGGCGGCGGAGGAGG----------AGGAGT------------TGCAGCA-------------------------------------------------------------- |
| droKik1 |
scf7180000302486:2317673-2317878 - |
|
TTCCGCATTAATCGTATCCGCATCGCAGCCGCACGAGGATTTCGCCGAAACGAGCGAGGATAT------------------------TTGTG---ACAAAGAGCCGAGGA--------------------CCGATGATAA-------------------CGGGG--GA--------------------GTC-----------------------CTAGACACAGAGGACCTTCACACGTCG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATAG---------ACGATAGCTCAGCAGCAGCAACA--------------------------------------------------------GTCGA--------GGAGGACGCAGCCAACGACGC---------GGAAGT------------CGAGGCAG-TCATCATGG--------------------------------------------------- |
| droFic1 |
scf7180000453342:466821-467010 - |
|
AGCAGCAGCAGCAGAAGAAGC--A------------------------------------------GCAA---------CAACCGC-----------------------------------------------------------------------------------ATCAGCAACAGCAACAGCAACCCGAAGCG-----------------------------------------------------------------------------------------------------------------------------------------------------------------------GAAACGGAAATAGACA--------------------------------------CAGAGCCAGAGAGC---------G-AAACGGAGACCGAGACGGA----------------------------------------------------------------------AACGGAGACC----------------GAGACGG-AGACGGAGAGAGA-----G--------------------------------------------------------ACCGA--------GAGGGAGAGGGAGACAGAAAC---GGAAGCGGAAAC------------AGAAACCG---------A--------------------------------------------------- |
| droEle1 |
scf7180000490564:530855-531043 - |
|
AGCAGCTGCGGCAGAACGAGG-----------------GGGTTGCCGAGCCGAGCGCGGAAAATTCC---GATCAGGAGCACCAAATAAGTC---ACAAAGAGCCGAACAACG--------------GTATAGATGATAA-------------------GGAAA--CG--------------------CTC-----------------------CTAGAG---CAGGAGCTTCACGAATCAGC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A--------------------------------------------------------GTCGA--------GGAGGACGCAGCAATCGACGC---------GGAAGT------------CGAGGCAG-TCATCATGG--------------------------------------------------- |
| droRho1 |
scf7180000778255:59515-59679 - |
|
AGGGG-----------------------------------GTCGCCGAACCGAGTGCGGAAAATTCC---GACCAGGAGCACCAAATAAGTC---ACAAAGAGCCGAACA--------------------TCGATGATAA-------------------GGAAG--CG--------------------CTC-----------------------CTAGAG---CAGGACCTTAACAAATCAAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A--------------------------------------------------------GTCGA--------GGAGGACGCAGCTATCGACGC---------GGAAGT------------CGAAGCAG-TCATCATGG--------------------------------------------------- |
| droBia1 |
scf7180000302126:3411896-3412106 - |
|
AGCAGCAGCAGCAGCAGCATC--A------------------------------------------GCAGGGCAACGGGCAGTGACCTCACGCCCACGGACCTG-C---GACATGTGCTGCCCAGCAGCAACCACGATCA-------------------GCAGC--AG--------------------CAG-----------------------CA------------------------------------ACATCAGC---------------AGCAGCAGCA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAGCAGCAGCAACAGGAGCCCGCTGCCTAC-----------------------------------------------------------------GATGAC------------GAG-----------------------GACGA---GGA--GGAGGACGAGGACGACGAGGA---------TGAGGA------------CGAGGTGG-CCAGCA------------------------------------------------------ |
| droTak1 |
scf7180000415741:30333-30521 + |
|
AGCAGCAGCAGCTAAACGAGG-----------------GGGTCGCCGAGCCGAGAGCGGAAAATTCC---GATCAGGAGCAGCAAATAATAC---ACAAAGAGCCGAACATCG--------------ATATCGATGATAA-------------------GGAAG--CG--------------------CTC-----------------------CTAGAG---CAGGACCTTCGGAAACTAGC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A--------------------------------------------------------GTCGA--------GGAGGACGCAGCAACCGACGC---------GGAAGT------------CGAGGCAG-TCATCATGG--------------------------------------------------- |
| droEug1 |
scf7180000409711:1599910-1600041 - |
|
AGGAGCAACAAA--------------------------------------------------------------------------TAAGTC---ACAAAGAGCCGAAGA-----------------ACATCGATGATAA-------------------GGATA--CG--------------------CTC-----------------------CTAGAC---CAGGAACTTCAGCAATCATC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A--------------------------------------------------------GTTGA--------GGAGGACGCAGCAAACGACGC---------GGAAGT------------CGAGGCAG-TCATCATGG--------------------------------------------------- |
| dm3 |
chr3L:9743944-9744145 - |
|
GCCGAGTGCGGAAAAT------------------------------------------------TCG---GATCAGGAGCAGGCACTTTGTGACTACAAAGAGCCGAACA--------------------TTGATGATAA-------------------GGAAG--CG--------------------CTT-----------------------ATAGAG---CAGGACCTTCACAAATCATC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A--------------------------------------------------------GTCGA--------GGAGGACGCAGCTAACGACGC---------GGAAGT------------CGAGGCAG-TCATCATGG-TACCACA-ATAAAATATACAATTTTGAATAGTCGTTCG--GAGTTTCGTT |
| droSim2 |
3r:7028489-7028700 + |
|
AGCAGCAGCAGCAGCAGCAAC--A------------------------------------------GCAG---------CAGTCAC---C---------------CATCA--------------------GC-AGCATGA-------------------GCAACGG-----------------CAACATGCGGCAGCA------------------------------------------------------ACAGCAGC---------------AGCAGCAGCAACATCGACGCCAGCAGCAGCT-----------------------------------------------------------------------------------TCCGCCTGC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAGAC------------GATCATCATCGATGAGAGCGACA---A-TGA---GCTGGAGGAGGAGCAGCAGATGAGGC---------TGAGCTTTGAGCGGTACATGGAGCAG-CTGAAGCAG--------------------------------------------------- |
| droSec2 |
scaffold_0:1977542-1977707 - |
|
AGCAGGCACT-----------------------------------------------------------------------------TTGTGACTACAAAGAGCCGAACA--------------------TCGATGATAA-------------------GGAAG--CA--------------------CTC-----------------------CTCGAG---CAGGACCTTCACAAATCATC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A--------------------------------------------------------GTCGA--------GGAGGACGCAGCCAACGACGC---------GGAAGT------------CGAGGCAG-TCATCATGG-TACCGCA-AGAAAATATACAATTTTGAATAGCCATTCG------------ |
| droYak3 |
2R:15025622-15025833 + |
|
AGCAACAGCAGCAACAGCAAC--A------------------------------------------GCAG---------CAAGCGC---T---------------G--------------------------------------------------------------GAGTTGCAACAGCAACAAG-----------CAATTAATGCAAATGCGGAGA-GCA----------------------------------------------GCCGGC---------------------------AGCCAAATCGAGAAGGAGGCAGGCCAAGCAC----ATCGAGA-TCAAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCG-AATGCCTGGAGAATGT----CGG-----AGCTGCAGC-----AGGAGCAGAAGCA--------------------------------------------GCAGC--------AGAAGCAGCAGCAGTAGAAGC---------AGCAAC------------AGCAGCAG---------T--------------------------------------------------- |
| droEre2 |
scaffold_4929:3383204-3383445 - |
|
AGCAGCAACAGCAGCAGCAGC--A------------------------------------------GCAA---------CAACAGC-----------------------------------------------------------------------------------AACAACAGCAGCA----------------------------------------------------------------------ACAGCAGC---------------ATCCGCAGCA------------------ACACTACC-------------------AGC----T---------------------------AAACAGGAAACAGTTGCAGTTGCTGCCGCCTGTGTTGCTCCTCGCAAATTGGCAAAAATTCAGAAAAT---------TCCGA------------AGGA----------------------------------------------------------------------ACCCGAG-------------------GAGCTGA-AAGCCAAGAAAAAGGCAAA--------------------------------GTGT-----------------------GA--------GGCAGCAGCAGCAGCAGTAGC---------AGCAGC------------GGCGGCGACTCATTACGA-AAAAA--------------------------------------------- |