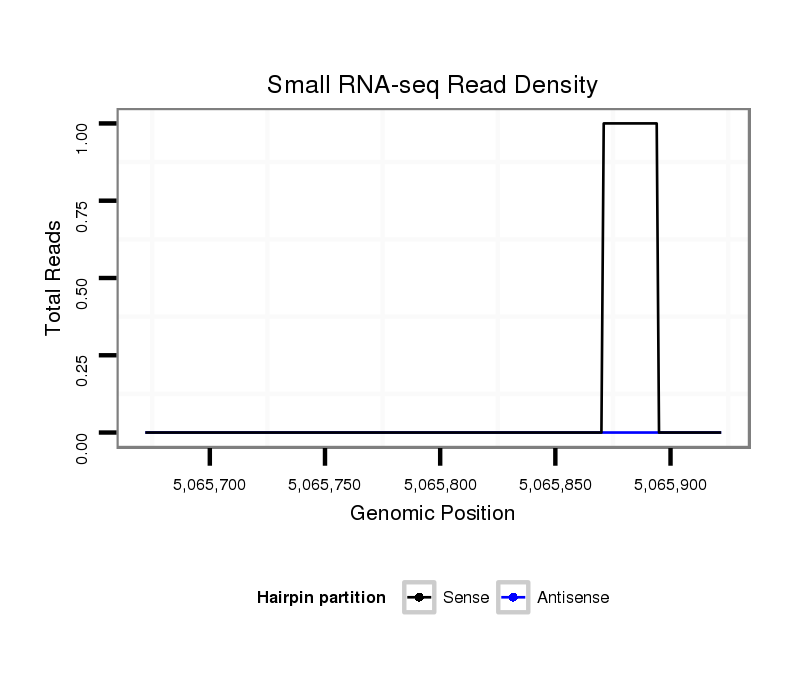
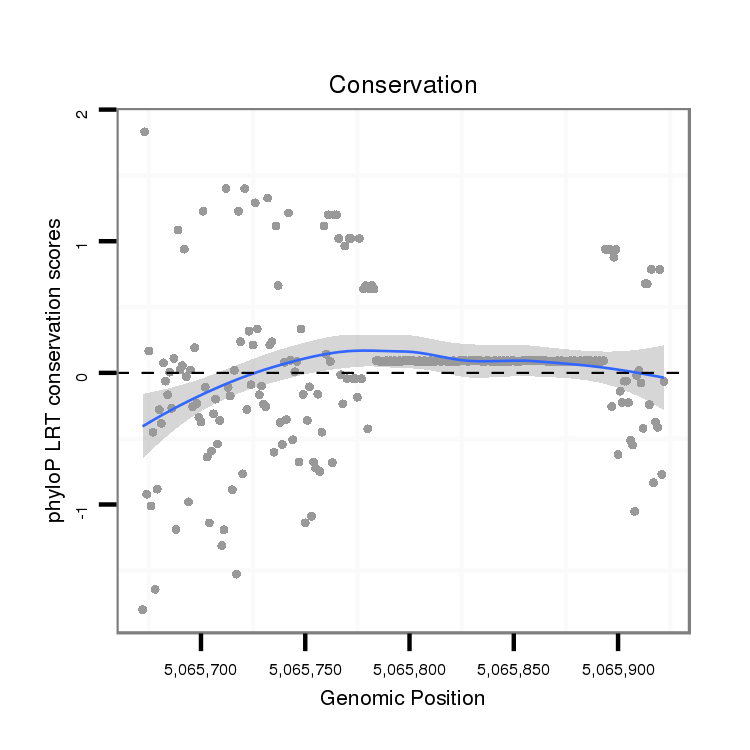
| droPer2 |
scaffold_6:5065672-5065922 + |
dpe_1929 |
ATAAGGCATAT----ACTT-CTCTCGCTCTCTGGG-CGTCTAAATA------TCGGTGCATTCA--CGCAAACATATCGATA---------------GATTTGTGTTGGTGTTGGTGGTTCAAAACGTTTTTCTTGAACTGAAGGCTGTCTCTGTTTTTTCCAGTGCAGCTAGTTCGCTCCATAAGAATTTTGGATCCAGTGACTCATGGCTATTTTTTTTTACCGGCAGACGGACAGCAGAAAAGTGTGATTTACACGAAAATTGAAACATCCAAAGAC |
| dp5 |
2:29692009-29692263 + |
|
CTAAGGCATATACATACTT-CTCTCGCTCTCTGGG-CGTCTAAATA------TCGGTGCATTCA--CGCAAACATATCGATA---------------GATTTGTGTTGGTGTTGGTGGTTCAAAACGTTTTTCTTGAACTGAAGGCTGTCTCTGTTTTTTCCAGTGCAGCTAGTTCGCTCCATAAGAATTTTGGATCCAGTGACTCATGGCTATTTTTTTTTACCGGCAGACGGACAGCAGAAAAGTGTGATTTACACGAAAATTGAAACATCCAAAGAC |
| droWil2 |
scf2_1100000004968:210150-210250 - |
|
ATTATGCACCA------TC-CCC------ATATAGTTTCTCGCTGG------TTA--CCAGTGGGAAGCAGGTATATCG-------------TAATTGTTTAGTATTTTAGTAGTAGATTTAAAGGGTTTTTTTT------------------------------------------------------------------------------------------------------------------------------------------------- |
| droBip1 |
scf7180000395339:70164-70174 - |
|
TTTAAGAAAAT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droKik1 |
scf7180000298139:19592-19704 - |
|
ATTAACTCATT------------TTGAGCTTTGTG-CATTTGAATGCGATTTTTTTTCCACTCA--TGCAGATGTCTAG--TGTG------------TATACACTTTCGTAGTGATGGTCAAA--------------------------------------------------------------------------------------------------------------------------------TTTTCAGAGTTGAACCGAA---------- |
| droEle1 |
scf7180000491278:2008876-2008980 - |
|
ATAGATAGAGTGCACGAGTCCA--------------CATTTAATAA------TCGATG--TTAA--TACAGATGTACAGACA--GGCGAACATGG----------TCGATCAAGAGGGTTTAAAACGCTTCCTTCGACCTG------------------------------------------------------------------------------------------------------------------------------------------- |
| droRho1 |
scf7180000777559:12989-13068 - |
|
ATAAGTCCGAA----A--T-CTCTTGCTCATATAGTTTCCCAGATC------TCG--ACGTTCA--TACGGACG--------------GACATGG----------CAGAAAAAT---------------------------------------------------------------------------------------------------------------------------------------------------------------CGGTGTT |
| droBia1 |
scf7180000302244:163606-163633 - |
|
TTATACGAAAT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAAAAGTATTCTAAGCT |
| droTak1 |
scf7180000414330:4558-4642 + |
|
ATTAGCGAAATTTACAATC-GCCATGCATTTAGAG-CAGC----GG------TCGGCACACTCA--TACAATTATATT--TT---------------GTTTTATATTTGTGTGAGT-------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droEug1 |
scf7180000409804:1505986-1506008 + |
|
GTGA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTACACGATTAAGCAAGT---------- |