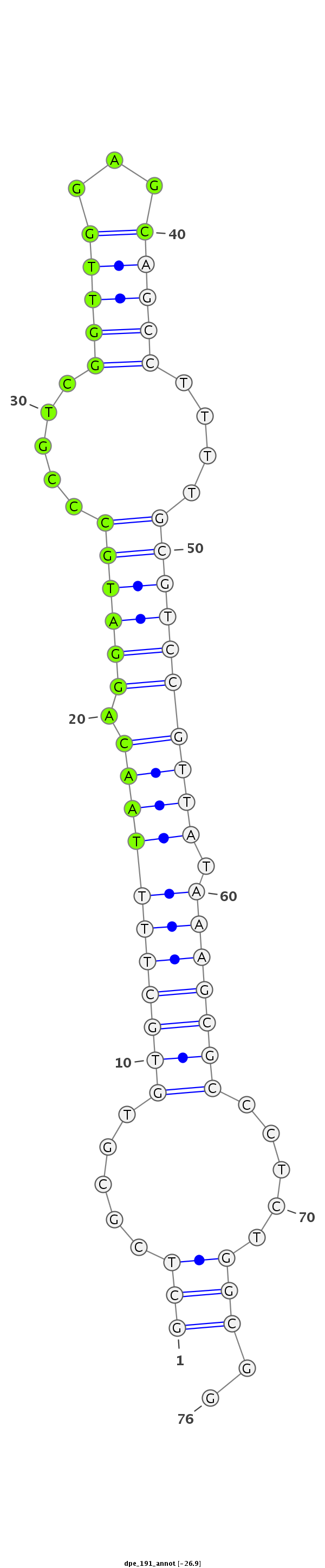
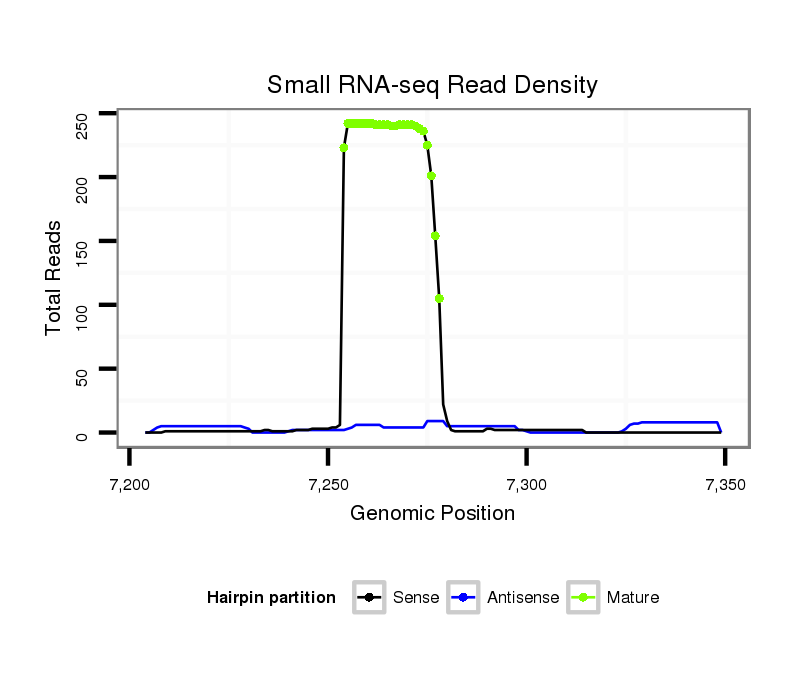
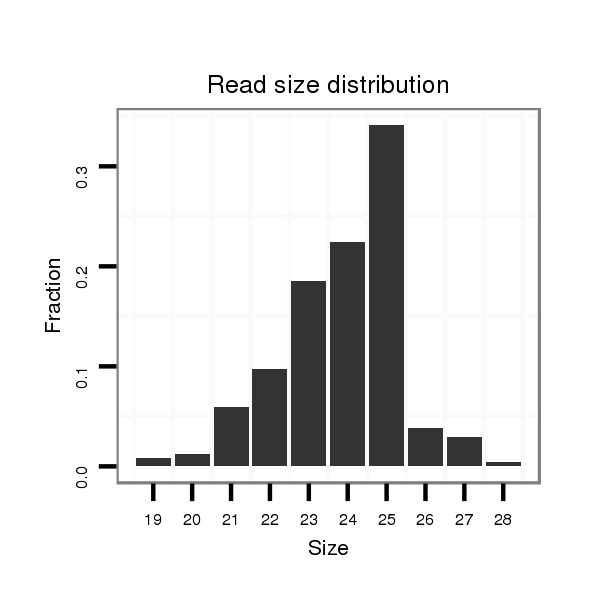
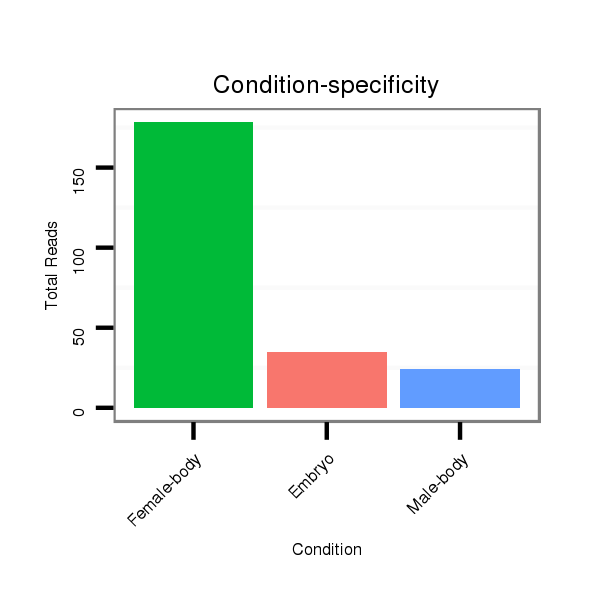
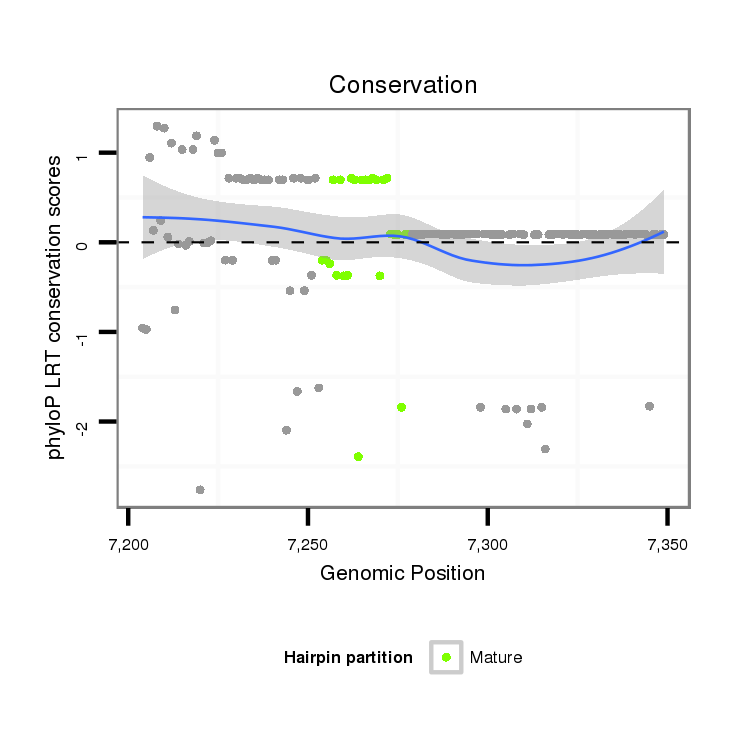
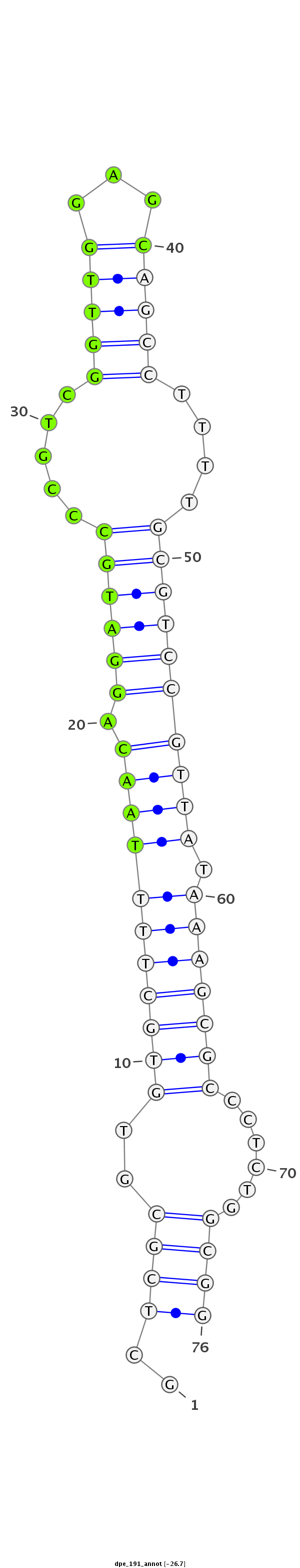
ID:dpe_191 |
Coordinate:scaffold_792:7254-7299 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -26.7 | -26.4 | -25.9 |
 |
 |
 |
intergenic
No Repeatable elements found
|
CAGGTTGCTAGCCTGCCATTTCCCTTTTGCTGTCCGCTCGCGTGTGCTTTTAACAGGATGCCCGTCGGTTGGAGCAGCCTTTTGCGTCCGTTATAAAGCGCCCTCTGGCGGAGGGTAGTCGCCTGGCGGCTCCGGTAGATCTGCGC
***********************************(((.....(((((((((((.((((((.....(((((...)))))....)))))))))).))))))).....)))..*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M042 female body |
M021 embryo |
V111 male body |
V042 embryo |
V057 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................TAACAGGATGCCCGTCGGTTGGAGC....................................................................... | 25 | 0 | 1 | 78.00 | 78 | 53 | 8 | 10 | 7 | 0 |
| ..................................................TAACAGGATGCCCGTCGGTTGGAG........................................................................ | 24 | 0 | 1 | 47.00 | 47 | 44 | 1 | 2 | 0 | 0 |
| ..................................................TAACAGGATGCCCGTCGGTTGGA......................................................................... | 23 | 0 | 1 | 42.00 | 42 | 22 | 13 | 4 | 3 | 0 |
| ..................................................TAACAGGATGCCCGTCGGTTGG.......................................................................... | 22 | 0 | 1 | 19.00 | 19 | 16 | 3 | 0 | 0 | 0 |
| ..................................................TAACAGGATGCCCGTCGGTTG........................................................................... | 21 | 0 | 1 | 10.00 | 10 | 10 | 0 | 0 | 0 | 0 |
| ..................................................TAACAGGATGCCCGTCGGTTGGAGCA...................................................................... | 26 | 0 | 1 | 9.00 | 9 | 7 | 0 | 2 | 0 | 0 |
| ..................................................TAACAGGATGCCCGTCGGTTGGAGCAG..................................................................... | 27 | 0 | 1 | 7.00 | 7 | 1 | 0 | 6 | 0 | 0 |
| ..................................................TAACAGGATGCCCGTCGGTTGGAGT....................................................................... | 25 | 1 | 1 | 5.00 | 5 | 2 | 3 | 0 | 0 | 0 |
| ...................................................AACAGGATGCCCGTCGGTTGGAGC....................................................................... | 24 | 0 | 1 | 5.00 | 5 | 5 | 0 | 0 | 0 | 0 |
| ...................................................AACAGGATGCCCGTCGGTTGG.......................................................................... | 21 | 0 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 |
| ...................................................AACAGGATGCCCGTCGGTTGGA......................................................................... | 22 | 0 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 |
| ...................................................AACAGGATGCCCGTCGGTTGGAGCA...................................................................... | 25 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 |
| ......................................................................................TCCGTTATAAAGCGCCCTCTGGCGG................................... | 25 | 0 | 4 | 2.50 | 10 | 10 | 0 | 0 | 0 | 0 |
| ..................................................TAACAGGATGCCCGTCGGTT............................................................................ | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..................................................TAACAGGATGCCCGTCGGTTGA.......................................................................... | 22 | 1 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 |
| ...................................................AACAGGATGCCCGTCGGTTGGAG........................................................................ | 23 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..................................................TAACAGGATGCCCGTCGGT............................................................................. | 19 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..................................................TAACAGGATGCCCGTCGGTTGGAA........................................................................ | 24 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..................................................TAACAGGATGCCCGTCGGTTGGAAA....................................................................... | 25 | 2 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 |
| ..................................................TAACAGGATGCCCGTCGGTTGGAGCGA..................................................................... | 27 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TAACAGGAGGCCCGTCGGTTGGAGC....................................................................... | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TAACGGGAGGCCCGTCGGTTGGA......................................................................... | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TAACAGGACGCCCGTCGGTTGGA......................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TAACAGGATGCCCGTCGGTTGGAGCAGC.................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TAACAGGAGGCCCGTCGGTTG........................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TAACAGGATCCCCGTCGGTTGGAGCA...................................................................... | 26 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....TGCTAGCCTGCCATTTCCCTTTTGCTG.................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TAACAGGATGCCCGTCTGTTGGAGC....................................................................... | 25 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................TTTTAACAGGATGCCCGTCGGTTGGA......................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TAACAGGAGGCCCGTCGGTTGGAGCTA..................................................................... | 27 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................CAGGATGCCCGTCTGTTGGAG........................................................................ | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................GAACAGGATGCCCGTCGGTTGGAGC....................................................................... | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TAACAGGATGCCCGTCGGTTGGAAC....................................................................... | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................TGTGCTTTTAACAGGATGCCCGTCGG.............................................................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................AACAGGATGCCCGTCGGTTGGAA........................................................................ | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................TCGGTTGGAGCAGCCTTTTGCGTC.......................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TAACAGGATGCCCGTCGGTA............................................................................ | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................TTAACAGGATGCCCGTCGGTTGGAGCA...................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................CGCGTGTGCTTTTAACAGGATGCC.................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................TTAACAGGATGCCCGTCGGTTGG.......................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................TGTCCGCTCGCGTGTGCTTTTAACAGGA........................................................................................ | 28 | 0 | 2 | 1.00 | 2 | 1 | 1 | 0 | 0 | 0 |
| ...................................................AACAGGATGCCCGTCGGTTG........................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................TCCGTTATAAAGCGCCCTCTGGC..................................... | 23 | 0 | 4 | 0.75 | 3 | 3 | 0 | 0 | 0 | 0 |
| ......................................................................................TCCGTTATAAAGCGCCCTCTGGCGGAGG................................ | 28 | 0 | 4 | 0.75 | 3 | 3 | 0 | 0 | 0 | 0 |
| ........................TTTTGCTGTCCGCTCGCGTGTGC................................................................................................... | 23 | 0 | 5 | 0.60 | 3 | 3 | 0 | 0 | 0 | 0 |
| ........................TTTTGCTGTCCGCTCGCGTGTG.................................................................................................... | 22 | 0 | 5 | 0.60 | 3 | 2 | 0 | 0 | 1 | 0 |
| ........................................................................................................GTGGCGGAGGGTAGTCGCC....................... | 19 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................GCTCGCGTGTGCTTTTAACAGGATG...................................................................................... | 25 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TAACAGGATGCCCGTCGGTTGGG......................................................................... | 23 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TAAAAGGATGACCGTCGGCTGGAG........................................................................ | 24 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................GCGGAGGGTAGTCGCCTGGCGG................. | 22 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................GTCCGTTATAAAGCGCCCTCTGGCGG................................... | 26 | 0 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...................................GCTCGCGTGTGCATTTAACAGGATG...................................................................................... | 25 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................TCTGGCGGAGGGTAGTCGCCT...................... | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................TGGCGGCTCCGGTAGATCTGCGC | 23 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................TCCCTTTTGCTGTCCGCTCGCGTG...................................................................................................... | 24 | 0 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..........................................................TGCCCGTCGGTTGGAGCAGCCTTT................................................................ | 24 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................CTGGCGGCTCCGGTAGATCTGC.. | 22 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................TCCGTTATCAAGCGCCCTCTGGCGG................................... | 25 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................TGCGTCCGTTATAAAGCGCCCTCTGGCGG................................... | 29 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................TCCGTTATAAAACGCCCTCTGGCGG................................... | 25 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................CCGTTATAAAGCGCCCTCTGGCGG................................... | 24 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................GCGTCCGTTATAAAGCGCCCTCT........................................ | 23 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................TTTTGCGTCCGTTATAAAGCGCCCT.......................................... | 25 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................TCCGTTATAAAGCGCCCTCTGGCCGAGG................................ | 28 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................TCCGTTATAAAGCGCCCTCTGGCGGA.................................. | 26 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................TTTGCGTCTGTTATAAAGCGCCCTC......................................... | 25 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................TTTGCGTCCGTTATAAAGCGCCCTCTG....................................... | 27 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................TATAAAGCGCCCTCTGGCGGAGG................................ | 23 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................TGCGTCCGTTATAAAGCGCCCTCTG....................................... | 25 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................CCGTTATAAAGCGCCCTCTG....................................... | 20 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................TTTGCGTCCGTTATAAAGCGCCCTCT........................................ | 26 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................GTCCGTTATAAAGCGCCCTCTGG...................................... | 23 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................GTCCGTTATAAAGCGCCCTCTGGC..................................... | 24 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................TTTGCTGTCCGCTCGCGTG...................................................................................................... | 19 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................TTCCCTTTTGCTGTCCGCTCGC......................................................................................................... | 22 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............TGCCATTTCCCTTTTGCTGTCCGCT............................................................................................................ | 25 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................TTCCCTTTTGCTGTCCGCTCGCGTG...................................................................................................... | 25 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................TTTGCTGTCCGCTCGCGTGTGCTT................................................................................................. | 24 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............TGCCATTTCCCTTTTGCTGTCCGCTC........................................................................................................... | 26 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................TTTCCCTTGTGCTGTCCGCTCG.......................................................................................................... | 22 | 1 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................TCTGGCGGAGGGTAGTCA......................... | 18 | 1 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............TGCCATTTCCCTTTTGCTGTCCGCTCG.......................................................................................................... | 27 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............TGCCATTTCCCTTTTGCTGTCCGC............................................................................................................. | 24 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................TTCCCTTTGGCTGTCCGCTCGCGTA...................................................................................................... | 25 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................GTGGGTAGTCGGCGGGCGGC................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
|
GTCCAACGATCGGACGGTAAAGGGAAAACGACAGGCGAGCGCACACGAAAATTGTCCTACGGGCAGCCAACCTCGTCGGAAAACGCAGGCAATATTTCGCGGGAGACCGCCTCCCATCAGCGGACCGCCGAGGCCATCTAGACGCG
***********************************(((.....(((((((((((.((((((.....(((((...)))))....)))))))))).))))))).....)))..*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M042 female body |
M021 embryo |
V042 embryo |
V111 male body |
V057 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................GACCGCCGAGGCCATCTAGACGC. | 23 | 0 | 3 | 3.33 | 10 | 10 | 0 | 0 | 0 | 0 |
| .......................................................................CTCGTCGGAAAACGCAGGCAATA.................................................... | 23 | 0 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 | 0 |
| .........................................................................................................................GGACCGCCGAGGCCATCTAGACGC. | 24 | 0 | 3 | 2.33 | 7 | 6 | 0 | 0 | 1 | 0 |
| ..CCAACGATCGGACGGTAAAGGGAAA....................................................................................................................... | 25 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .....................................................GTCCTACGGGCAGCCAACCTCGT...................................................................... | 23 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .....................................AGCGCACACGAAAATTGTCCTAC...................................................................................... | 23 | 0 | 2 | 1.50 | 3 | 3 | 0 | 0 | 0 | 0 |
| ........................................................................................................................CGGACCGCCGAGGCCATCTAGACGC. | 25 | 0 | 3 | 1.33 | 4 | 4 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CGCCGAGGCCATCTAGACGC. | 20 | 0 | 4 | 1.25 | 5 | 3 | 2 | 0 | 0 | 0 |
| ....................................................TGTCCTACGGGCAGCCAACCTCGT...................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .ACCAACGATCGGACGGTAAAGGGAA........................................................................................................................ | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................CTCGTCGGAAAACGCAGGCAATATT.................................................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................TTGTCCTACGGGCAGCCAACCTCGT...................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...CAACGATCGGACGGTAAAGGGAA........................................................................................................................ | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...CAACGATCGGACGGTAAAGGGAAA....................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................ACCGCCGAGGCCATCTAGACGC. | 22 | 0 | 4 | 1.00 | 4 | 4 | 0 | 0 | 0 | 0 |
| ....................................GAGCGCACACGAAAATTGTCCTAC...................................................................................... | 24 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .......................................................................CTCGTCGGAAAACGCAGGCAATATTT................................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....AACGATCGGACGGTAAAGGGA......................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................CCGCCGAGGCCATCTAGACGC. | 21 | 0 | 4 | 0.75 | 3 | 1 | 2 | 0 | 0 | 0 |
| .........................................................................................................................GGACCGCCGAGGCCATCTAGACG.. | 23 | 0 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..........CGGACGGTAAAGGGAAAACGACAG................................................................................................................ | 24 | 0 | 5 | 0.60 | 3 | 2 | 0 | 0 | 1 | 0 |
| ..GCAACGATCGGACGATAC.............................................................................................................................. | 18 | 3 | 20 | 0.60 | 12 | 5 | 0 | 1 | 0 | 6 |
| .................................GGCGAGCGCACACGAAAATTG............................................................................................ | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................ACGACAGGCGAGCGCACACGAAAAT.............................................................................................. | 25 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................CGAGCGCACACGAAAATTGTCCTAC...................................................................................... | 25 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................GCGCACACGAAAATTGTCCTAC...................................................................................... | 22 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................CAGCGGACCGCCGAGGCCATCTAGACGC. | 28 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................GGACCGCCGAGGCCTTCTAGACG.. | 23 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................GACCGCCGAGGCCAACTAGACGC. | 23 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................GGACCGCCGAGGCCAGCTAGACGC. | 24 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................CCCATCAGCGGACCGCCGAGGCCAT......... | 25 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................GACCGCCGAGGCCATCTAGAC... | 21 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................CGGACCGCCGAGGCCATCTAGACG.. | 24 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................CGGACCGCCGAGGCCATCTA...... | 20 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CGCCGAGGCCATCTAGACG.. | 19 | 0 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................AGGCAATATTTCGCGGGAGACC...................................... | 22 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........CGGACGGTAAAGGGAAAACGACAGG............................................................................................................... | 25 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............ACGGTAAAGGGAAAACGACAGGC.............................................................................................................. | 23 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........CGGACGGTAAAGGGAAAACGACA................................................................................................................. | 23 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........GGACGGTAAAGGGAAAACGACAGG............................................................................................................... | 24 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................ATTTCGCAGGAGACAGCCTT................................. | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................GACCACCGGGGCTATCTAGA.... | 20 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................CGACAGGCGGGGGCACAC.................................................................................................... | 18 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 |
| .TCCAACAAAAGGACGGTAAAG............................................................................................................................ | 21 | 3 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..CCAACGATCGGACCATAC.............................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droPer2 | scaffold_792:7204-7349 + | dpe_191 | CAGGTTGCTAGCCTGCCATTTCCCTTTTGCTGTCCGCTCGCGTGTGCTTTTAACAGGATGCCCGTCGGTTGGAGCAGCCTTT-------------TGCGTCCGTTATAAAGCGCCCTCTGGCGGAGGGTAGTCGCCTGGCGGCTCCGGTAGATCTGCGC |
| dp5 | Unknown_group_452:9048-9206 + | CAGGTTGGTAGCCTGCTAATTCCCTTTTGCTGTCCGCTCGAGTATGCTTCTAACAGGATGGCCGTCGGTTGGGGCAGCCTTTTGCCCAAGCCATTTGCGTCCGTTATGAAGCGCTCTTTGTTGGGCGGTAGTCGCCTGGCGGCTCCGGTAGATCCGCGC | |
| droWil2 | scf2_1100000003774:267-273 - | CCGGTTG-------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droVir3 | scaffold_682:680-695 + | GAGTTTGCTTGCTTGC----------------------------------------------------------------------------------------------------------------------------------------------- | |
| droMoj3 | scaffold_6482:67751-67806 - | TG-------------CAATGTCCGTCTTGCTGTCCGTCCGTCTGTCCGTCCGTCCGTCTGTCCGTCTGT------------------------------------------------------------------------------------------ | |
| droAna3 | scaffold_13055:115393-115413 + | GT---CGCTTTCTCGCGTTTTCCC--------------------------------------------------------------------------------------------------------------------------------------- | |
| droBip1 | scf7180000393827:752-770 + | G--GTCGCTATCTCGCTATTT------------------------------------------------------------------------------------------------------------------------------------------ |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
Generated: 05/15/2015 at 02:39 PM