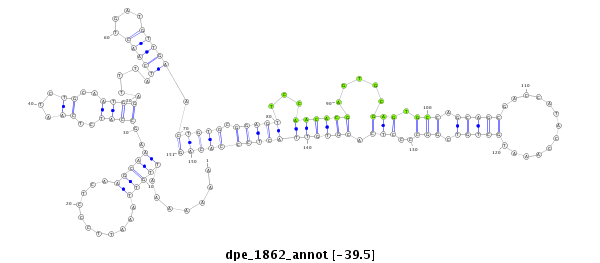
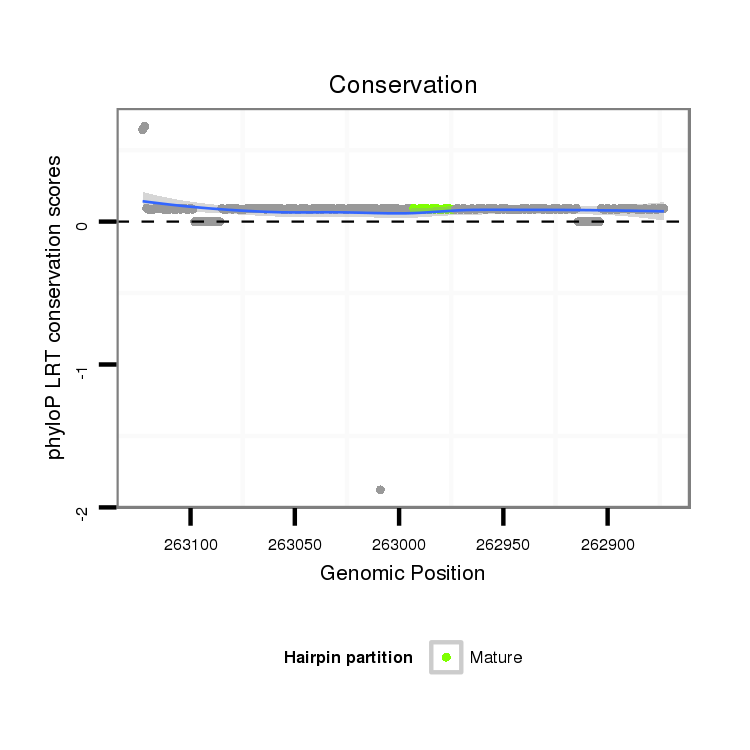
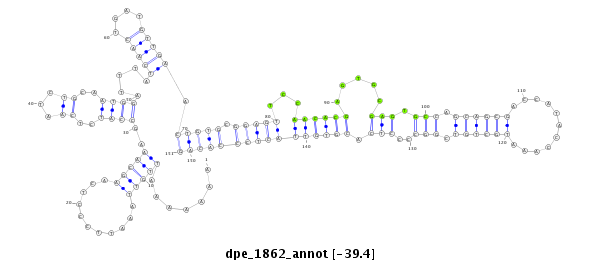
ID:dpe_1862 |
Coordinate:scaffold_59:262923-263073 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -39.4 | -39.0 | -38.9 |
 |
 |
 |
CDS [Dper\GL10998-cds]; exon [dper_GLEANR_1112:2]; intron [Dper\GL10998-in]
No Repeatable elements found
| mature | star |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CAAGGCAAAAGGCCTGCAGATCCACCACAAAGGGGAGGTCCAACATCGCCAAAAAAATTGTTAAATTCCCTCAAGCAAAGCCATCTCAATCTGCAATGGATTTATCAACTGATGTTGAACTGTGCGGAGTTCCAACACGAGTGCGAGTGCCAGCAGCGACCATACCAAATGCTGTCGGCCCCTCACGTGTTACTCCCACAGCACAAGCAGCCCCTCTGGTACCACCTGCTATTGAATCCCGACCCACCAAA **************************************************.......(((((...........)))))..((((..((...))..)))).....(((((....))))).(((((.(((((...((((((.....(((.(((.(((((.............))))).)))..))).))))))))))))))))************************************************** |
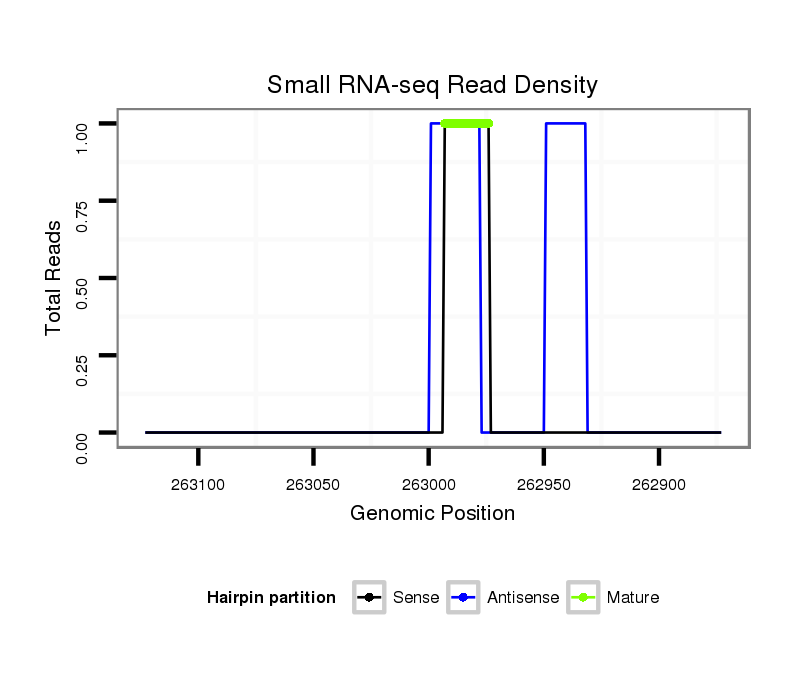
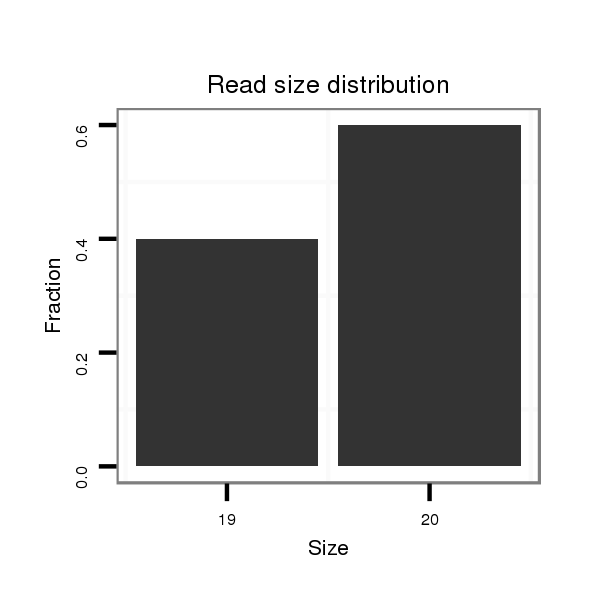
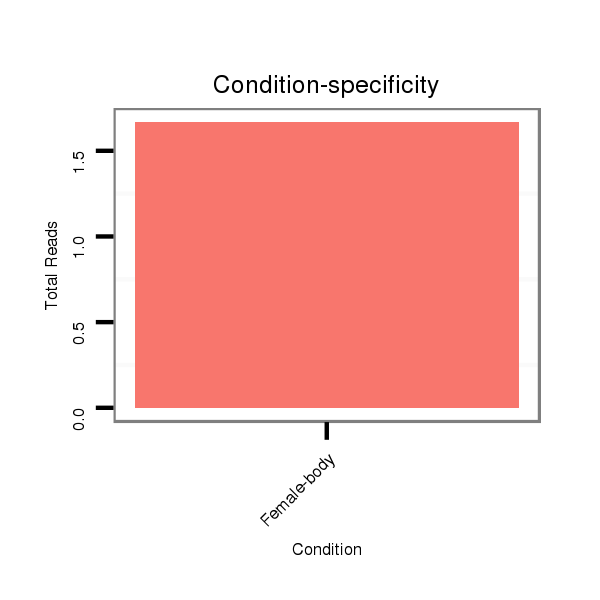
Read size | # Mismatch | Hit Count | Total Norm | Total | V057 head |
M042 female body |
M021 embryo |
|---|---|---|---|---|---|---|---|---|
| .............................AAGGGGAGGCCCTACATTGCCA........................................................................................................................................................................................................ | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..................................................................................................................................TCCAACACGAGTGCGAGTGC..................................................................................................... | 20 | 0 | 2 | 1.00 | 2 | 0 | 2 | 0 |
| ........................................................................................................................TGCGTGGAGTTCCAACACGGGT............................................................................................................. | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .................................................................................................................................TTCCAACACGAGTGCGAGT....................................................................................................... | 19 | 0 | 3 | 0.67 | 2 | 0 | 2 | 0 |
| ..........................................................................................................................................TTGTGCGAGTGCCAGCAGCG............................................................................................. | 20 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| ............................................................................................................CTGAGGTTGAAGTTTGCGGAG.......................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 |
| ..............................................................................................................GATGTTGACCTGTGCTGA........................................................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 |
| ...................................................AAAAAATTGTTAAATTCA...................................................................................................................................................................................... | 18 | 1 | 11 | 0.18 | 2 | 0 | 2 | 0 |
| ...............................................................................................................................................................................................................CAGCCCCTCTTCTACCAGCTGC...................... | 22 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 |
| .........................................................................................................................................CAAGTGCGCGTGCCAGCAGCT............................................................................................. | 21 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 |
| .........................................................................................................................................CGGGTGCGAGTGCCAGCGGCT............................................................................................. | 21 | 3 | 18 | 0.06 | 1 | 1 | 0 | 0 |
|
GTTCCGTTTTCCGGACGTCTAGGTGGTGTTTCCCCTCCAGGTTGTAGCGGTTTTTTTAACAATTTAAGGGAGTTCGTTTCGGTAGAGTTAGACGTTACCTAAATAGTTGACTACAACTTGACACGCCTCAAGGTTGTGCTCACGCTCACGGTCGTCGCTGGTATGGTTTACGACAGCCGGGGAGTGCACAATGAGGGTGTCGTGTTCGTCGGGGAGACCATGGTGGACGATAACTTAGGGCTGGGTGGTTT
**************************************************.......(((((...........)))))..((((..((...))..)))).....(((((....))))).(((((.(((((...((((((.....(((.(((.(((((.............))))).)))..))).))))))))))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M042 female body |
V111 male body |
V057 head |
|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................AGCCGGGGAGTGCACAAT........................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ............................................................................................................................GCCTCAAGGTTGTGCTCACGCT......................................................................................................... | 22 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 |
| ........................................................................................TAGACGTTACCTAGATAGTTGACT........................................................................................................................................... | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .....................................................................................................................................................GGTCGGCGCTGGTATGTTTTA................................................................................. | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 |
| ...................................................................................................................................................................................GGGAGTGCACAATGAGGGT..................................................... | 19 | 0 | 3 | 0.67 | 2 | 1 | 1 | 0 |
| .................................................................................................................................................................................CGGGGAGTGCACAATGAGGGT..................................................... | 21 | 0 | 3 | 0.67 | 2 | 0 | 2 | 0 |
| .................................................................................................................................AAGGTTGTGCTCACGCTCACG..................................................................................................... | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| ....................................................................................................................................GTTGTGCTCACGCTCACGGT................................................................................................... | 20 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 |
| ....................................................................................................................................................................................TGAGTGCACAATGAGGGTGT................................................... | 20 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 |
| ................................................................................................................................................................................CCGGGGAGTGCACAATGAGG....................................................... | 20 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ...........................................................................................................TGACTACTACTTGACACGCCTCAAGGTT.................................................................................................................... | 28 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ...............................................................................................................................................................................GCCGGGGAGTGCACAATGAGGG...................................................... | 22 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ................................................................................................................................................................................CCGGGGAGTGCACAATGAGGGTGT................................................... | 24 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 |
| .........................................................................................................GTTGACTACAACTTAGCACGCT............................................................................................................................ | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 |
| ..................................................................................................................TACTTGACACGCCTCAAGGTT.................................................................................................................... | 21 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ..................................................................................................................................................................................TGGGAGTGCATAATGAGGGTGT................................................... | 22 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 |
| ..............................................................................................................................................................................CGCCGGGGAGTGCACAAT........................................................... | 18 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ..................................................................................................................................................................................AAGGAGTGCACAATGAGGG...................................................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 |
| ....................................................................................................................................................................................GGAGTGCACAATGAGGGTGTCGTGT.............................................. | 25 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ...........................................................................................................TGACTACTACTTGACACGCCTCAAGGT..................................................................................................................... | 27 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| .............................................................................................................ACTACTACTTGACACGCCTCAAGGT..................................................................................................................... | 25 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droPer2 | scaffold_59:262873-263123 - | dpe_1862 | CAAGGCAAAAGGCCTGCAGATCCACCACAAAGGGGAGGT-----------CCAACATCGCCAAAA-AAATTGTTAAATTCCCTCAAGCAAAGCCATCTCAATCTGCAATGGATTTATCAACTGATGTTGAACTGTGCGGAGTTCCAACACGAGTGCGAGTGCCAGCAGCGACCATACCAAATGCTGTCGGCCCCTCACGTGTTACTCCCACAGCACAAGCAGCCCCTCTGGTACCACCTGCTATTGAATCCCGACCCACCAAA |
| dp5 | Unknown_group_388:3910-4148 + | CAAGGCAAAAGGCCTGCAGATCCAC-------------TCTCCCCTTTGTCCAACATCGCCAAAAAAAATTGTTAAATTCCCTCAAGCAAAGCCATCTCAATCTGCAATGGATTTATCAACTGATGATGAACTGTGCGGAGTTCCAACACGAGTGCGAGTGCCAGCAGCGACCATACCAAATGCTGTCGGCCCCTCACGTGTTACTCCCACAGCACAAGCA-----------ACCACCTGCTATTGAATCCCGACCCACCAAA | |
| droAna3 | scaffold_13045:901443-901444 + | CA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
Generated: 06/08/2015 at 04:49 PM