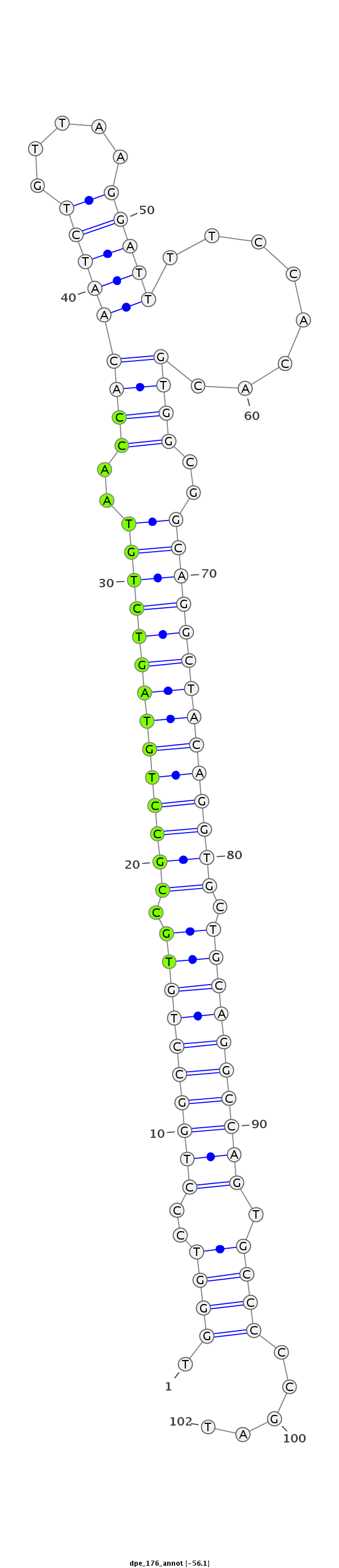
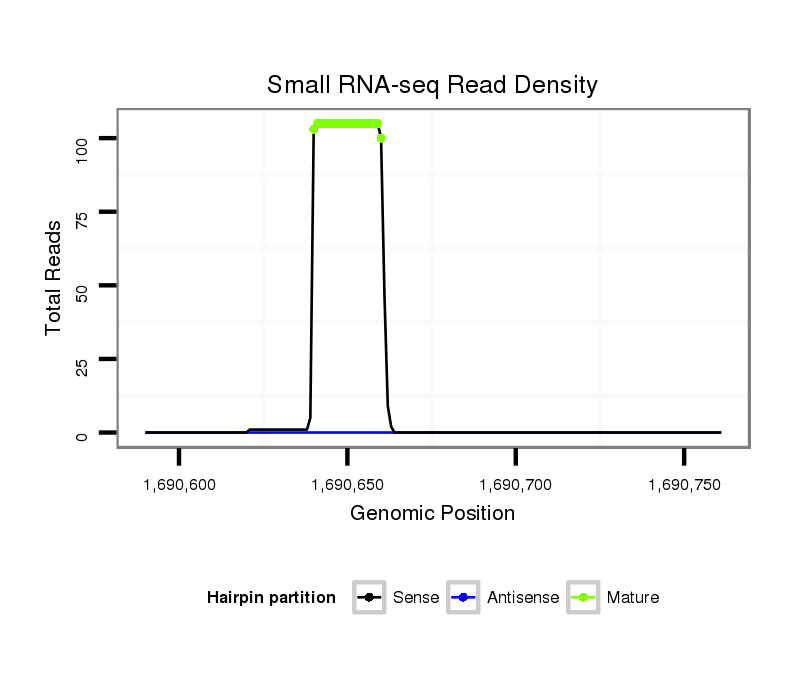
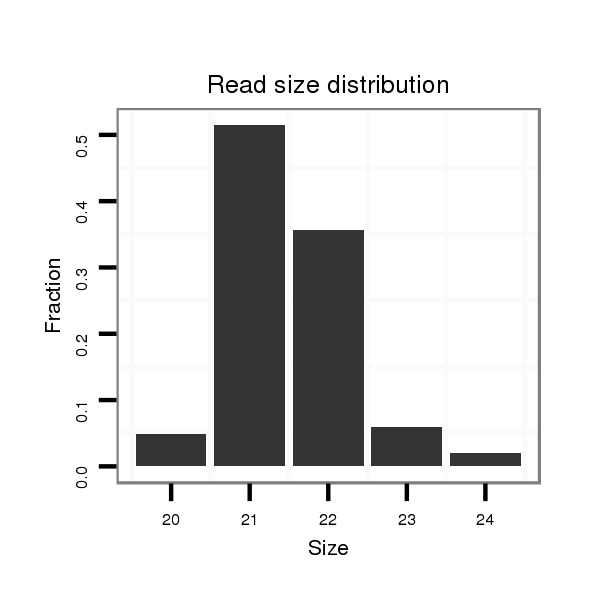

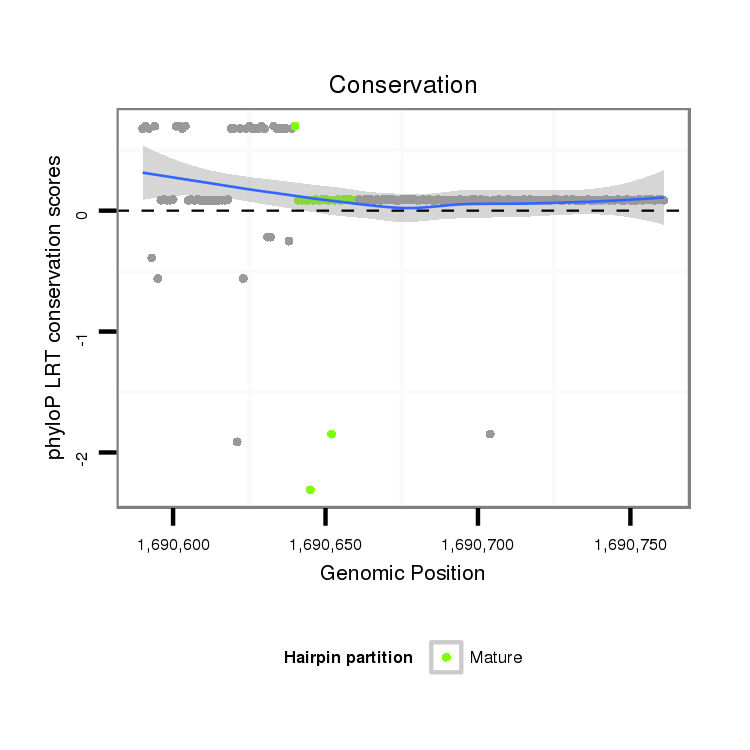
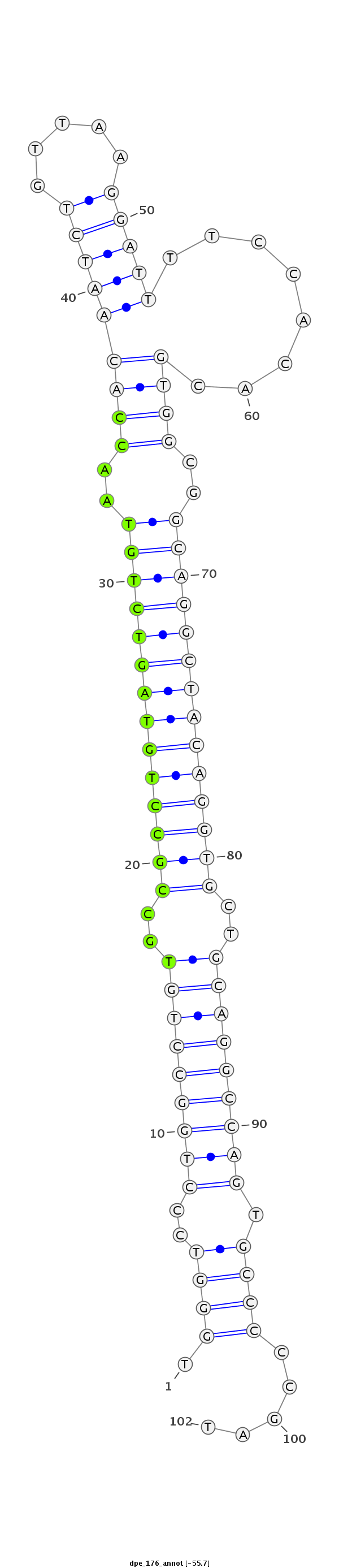
ID:dpe_176 |
Coordinate:scaffold_15:1690640-1690711 + |
Confidence:candidate |
Class:Canonical miRNA |
Genomic Locale:intron |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -55.7 | -55.5 | -55.1 |
 |
 |
 |
intron [Dper\GL16589-in]
No Repeatable elements found
|
CTCGTCCTGCAAACTGAGACGGGGCGGGAGGCCCGTGGGTCCCTGGCCTGTGCCGCCTGTAGTCTGTAACCACAATCTGTTAAGGATTTTCCACACGTGGCGGCAGGCTACAGGTGCTGCAGGCCAGTGCCCCCGATGCTCTCGCCACCGCCACCGTCGACGCTCAATCTAG
***********************************.((((..((((((((((.((((((((((((((..(((((((((.....)))))........))))..)))))))))))))).)))))))))).)))).....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M021 embryo |
M042 female body |
V057 head |
V042 embryo |
V050 head |
V111 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................TGCCGCCTGTAGTCTGTAACC..................................................................................................... | 21 | 0 | 1 | 50.00 | 50 | 31 | 9 | 5 | 1 | 3 | 1 |
| ..................................................TGCCGCCTGTAGTCTGTAACCA.................................................................................................... | 22 | 0 | 1 | 36.00 | 36 | 20 | 9 | 3 | 2 | 2 | 0 |
| ..................................................TGCCGCCTGTAGTCTGTAACCAC................................................................................................... | 23 | 0 | 1 | 6.00 | 6 | 3 | 2 | 1 | 0 | 0 | 0 |
| ..................................................TGCCGCCTGTAGTCTGTAAC...................................................................................................... | 20 | 0 | 1 | 5.00 | 5 | 0 | 3 | 2 | 0 | 0 | 0 |
| .................................................GTGCCGCCTGTAGTCTGTAACC..................................................................................................... | 22 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TGCCGCCTGTAGTCTGTAACCC.................................................................................................... | 22 | 1 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 | 0 | 0 |
| ..................................................TGCCGCCTGTAGTCTGTAACCACA.................................................................................................. | 24 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...................................................GCCGCCTGTAGTCTGTAACCA.................................................................................................... | 21 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ..................................................TGCCGCCTGTAGTCTGTCACCAC................................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TGCCGCCTGTAGGCTGTAACC..................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TGCCGCCTATAGTCTGTAACCA.................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TGCCGCCTGTAGTCTTTAAC...................................................................................................... | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................AGCCGCCTGTAGTCTGCAACCA.................................................................................................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TGCCGCCTGTAGACTGTAACCA.................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TGCCGCATGTGGGCTGTAACC..................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................GTGCCGCCTGTAGTCTGTAACCAC................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................CCCGTGGGTCCCTGGCCTG.......................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............GGGTCGGGTCGGGAGGCCCGTG....................................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TGCCGCCTGTAGACTGTAGCC..................................................................................................... | 21 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TGCCCCCTGTACTCTGAAACC..................................................................................................... | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................CACAGGTGCTGCATGCCTGT............................................ | 20 | 3 | 17 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
|
GAGCAGGACGTTTGACTCTGCCCCGCCCTCCGGGCACCCAGGGACCGGACACGGCGGACATCAGACATTGGTGTTAGACAATTCCTAAAAGGTGTGCACCGCCGTCCGATGTCCACGACGTCCGGTCACGGGGGCTACGAGAGCGGTGGCGGTGGCAGCTGCGAGTTAGATC
***********************************.((((..((((((((((.((((((((((((((..(((((((((.....)))))........))))..)))))))))))))).)))))))))).)))).....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M042 female body |
V057 head |
|---|---|---|---|---|---|---|---|
| ...................................................CGTCGGCCATCAGACATTG...................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 |
| ............................................CCGGACAAGGACGACATCAGAC.......................................................................................................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 1 |
| ........................................GGGACCAGACACGGTGGGC................................................................................................................. | 19 | 3 | 20 | 0.25 | 5 | 5 | 0 |
| ........................................GGGACCAGACACGGCGGGCGT............................................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 |
| .............................................CGGACGCGGTGGACACCAGA........................................................................................................... | 20 | 3 | 11 | 0.09 | 1 | 0 | 1 |
| .....................................CCGGGGACCGGACAAGGCGC................................................................................................................... | 20 | 3 | 16 | 0.06 | 1 | 0 | 1 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droPer2 | scaffold_15:1690590-1690761 + | dpe_176 | candidate | CTCGTCCTGCAAACTGAGACGGGGCGGGAGGCCCGTGGGTCCCTGGCCTGTGCCGCCTGTAGTCTGTAACCACAATCTGTTAAGGATTTTCCACACGTGGCGGCAGGCTACAGGTGCTGCAGGCCAGTGCCCCCGATGCTCTCGCCACCGCCACCGTCGACGCTCAATCTAG |
| dp5 | XL_group3a:1196731-1196902 - | dps_3582 | CTCGTCCTGCAAACTGAGACGGGGCGGGAGGACCGTGGGTCCCTGGCCTGTGCCGGCTGTAGCCTGTAACCACAATCTGTTAAGGATTTTCCACACGTGGCGGCAGGCTACAGGCGCTGCAGGCCAGTGCCCCCGATGCTCTCGCCACCGCCACCGTCGACGCTCAATCTAG | |
| droSec2 | scaffold_15:683877-683908 - | CTCTTG-----AACT--------------GGCCGGTGGGTCTTTGGCCAGT------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
Generated: 10/20/2015 at 06:50 PM