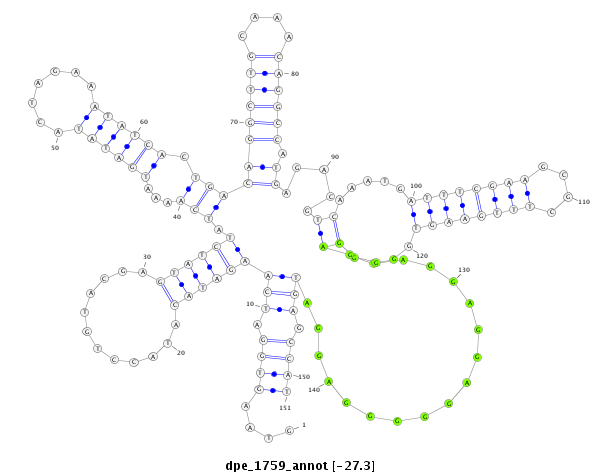
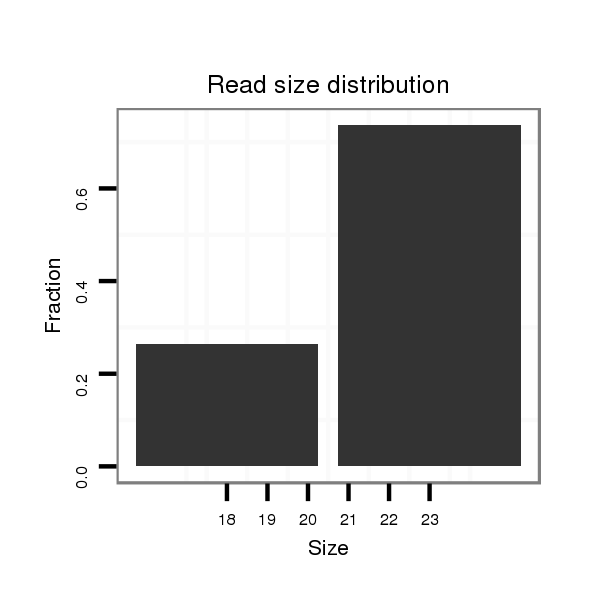
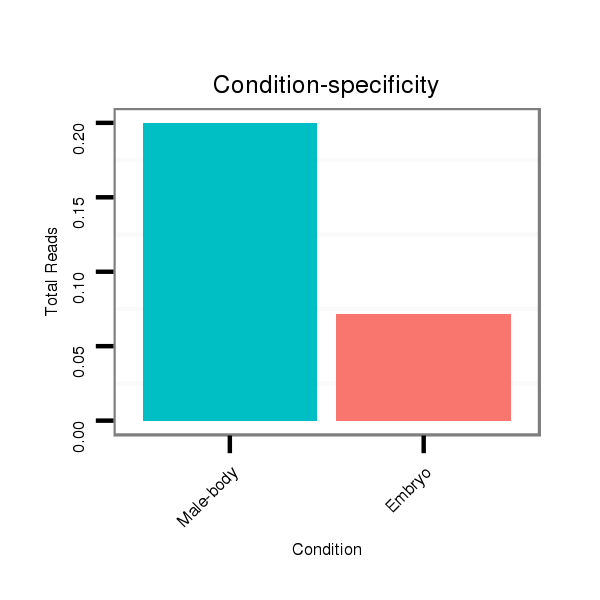
ID:dpe_1759 |
Coordinate:scaffold_5:2404639-2404789 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
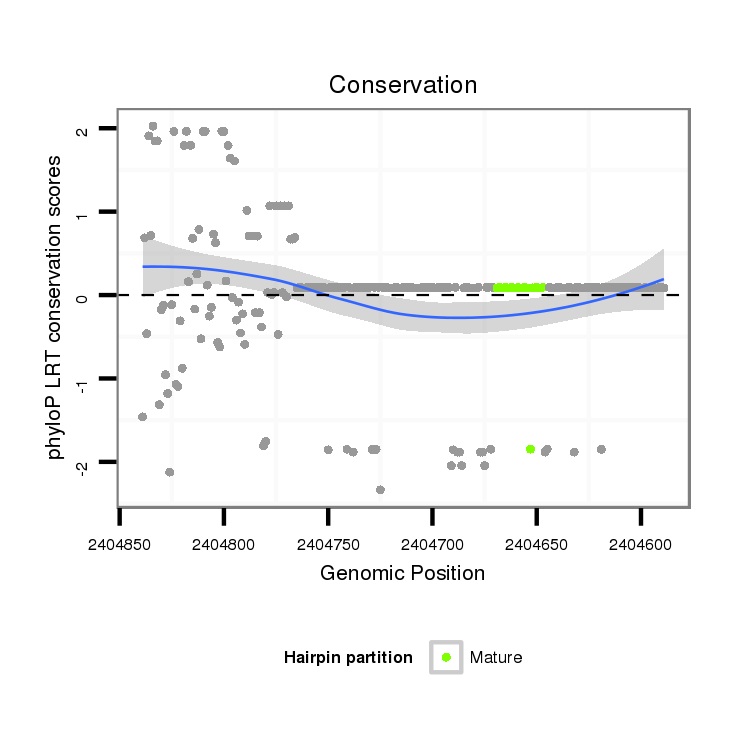
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
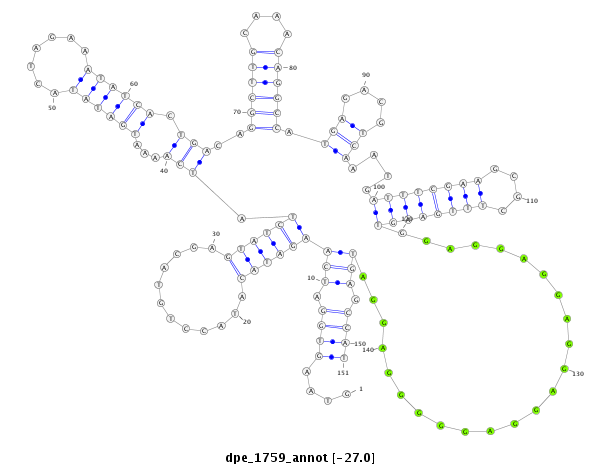
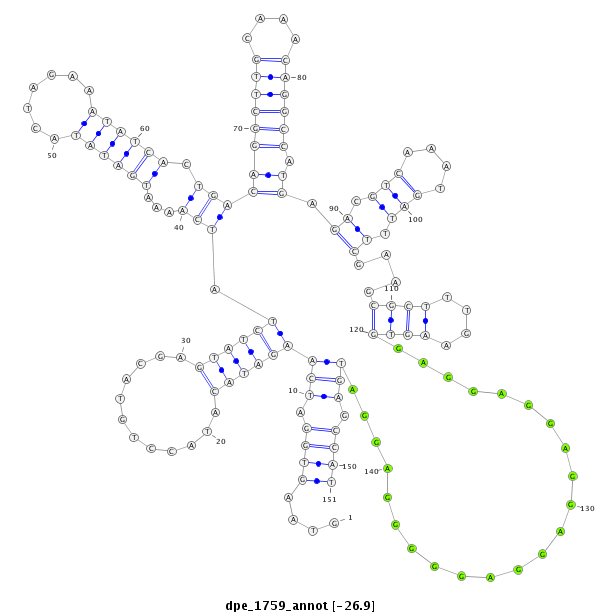
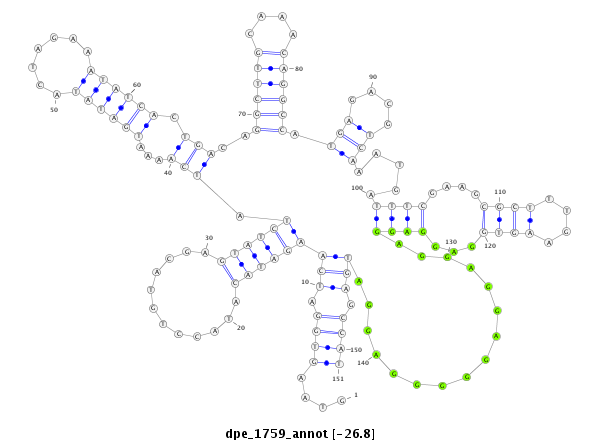
| -27.0 | -26.9 | -26.8 |
 |
 |
 |
exon [dper_GLEANR_8302:1]; CDS [Dper\GL25738-cds]; intron [Dper\GL25738-in]
| Name | Class | Family | Strand |
| (TCC)n | Simple_repeat | Simple_repeat | + |
| mature | star |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GGCCCTGGCGCCAACTTGGCGATGCGAACAAAACTCTTTACCGTCTTCACGTAAGTGGATCAAGATACATACCTGTACGAGTATCTATCAAAATGATATACTAGAAATATCACTGACAGGCTTGCAAACAGGCCATGAGACGTCAAATGATTTCGAAGCGCTTTGAAGTGGAGGAGGAGGAGGAGGGGGAGGATGAGCCATGCCTACTATCCGATTGTTCTATTTAGTGTGTTTTAATTCCACGATGGTTC **************************************************....((((.(((((((((............)))))).(((...((((((.......)))))).)))((((((((....)))))).)).....((.....((((((((....))))))))....))..................))).))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V111 male body |
M042 female body |
V050 head |
V042 embryo |
V057 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................GAGGAGGAGGAGGAGGGGGAGGA.......................................................... | 23 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................GAGGAAGAGGGGGAGGAT......................................................... | 18 | 1 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................GCTTTGAAGAGGATGAGGAGC....................................................................... | 21 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................ATAATTTGTAAGCGCTTTGAA.................................................................................... | 21 | 3 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................................AGTGGAGGAGGAGGAGGA................................................................... | 18 | 0 | 14 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................GAAGTGGAGGAGGAGGAGT.................................................................... | 19 | 1 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................GGAGGAAGAGGGGGAGGATA........................................................ | 20 | 2 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................CGGAGGAGGGGGGGGATGA....................................................... | 19 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................................................................................TTTGAAGAGGATGAGGAG........................................................................ | 18 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
|
CCGGGACCGCGGTTGAACCGCTACGCTTGTTTTGAGAAATGGCAGAAGTGCATTCACCTAGTTCTATGTATGGACATGCTCATAGATAGTTTTACTATATGATCTTTATAGTGACTGTCCGAACGTTTGTCCGGTACTCTGCAGTTTACTAAAGCTTCGCGAAACTTCACCTCCTCCTCCTCCTCCCCCTCCTACTCGGTACGGATGATAGGCTAACAAGATAAATCACACAAAATTAAGGTGCTACCAAG
**************************************************....((((.(((((((((............)))))).(((...((((((.......)))))).)))((((((((....)))))).)).....((.....((((((((....))))))))....))..................))).))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V057 head |
M042 female body |
|---|---|---|---|---|---|---|---|
| ......................................................................................................TCTTTATAGGGACTGTCATAAC............................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 |
| .................................................................................................................TCTGTCCGAACGTCTGTACGGT.................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 |
| ...................................................................................................................................................................................................................GCCAACAAGGTAAATCACAAAAAA................ | 24 | 3 | 2 | 0.50 | 1 | 1 | 0 |
| .........................................................................................................................................................................................................CGGATGTTAGGCTAACAAG............................... | 19 | 1 | 2 | 0.50 | 1 | 0 | 1 |
| ....................................................................................................................GTCCGGAAGTTTGACCGGTAC.................................................................................................................. | 21 | 3 | 9 | 0.11 | 1 | 1 | 0 |
| .................................................................................................................................................................................................................AGGCCCACAAGATAAACCA....................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droPer2 | scaffold_5:2404589-2404839 - | dpe_1759 | GGCCCTGGCGCCAACTTGGCGATGCGAACAAAACTCTTTACCGTCTTCACGTAAGTGGATCAAGATACATACCTGTACGAGTATCTATCAAAATGA-TATACTAGAAATATCACTGACAGGCTTGCAAACAGGCCATGAGACGTCAAATGATTTCGAAGCGCTTTGAAGTGGAGGAGGAGGAGGAGGGGGAGGATGAGCCATGCCTACTATCCGATTGTTCTATTTAGTGTGTTTTAATTCCACGATGGTTC |
| dp5 | 4_group1:2804354-2804604 + | GG-CCTGGCGCCGACTTGGCGATGCGAACAAAACTCTTTACCGTCTTCACGTAAGTGGTCCAAGATACATACCTGTACGAGTATCTATCGAAATGAATACACAAGAAATATTATTCACAGGCTTGCAAACAGGCCATGAGACGTCAAATTGTAAAGAAGCGCTAATAAATGGAGGAGGAGGAGGAGGAGGAGGAAAAGCCATGCCTACAATCCGATTGTTCCATTTAGTGTGTTTTAATTCCACGATGGTTC | |
| droWil2 | scf2_1100000007381:333-397 + | GTGCCTGGTGCAGTTTATCGGAAGATAAGAAGAATCGTTATCGTCGCATGG---------CAAGAAACAAACCT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droAna3 | scaffold_12916:969724-969767 + | CG-CCTGGTGCCGCTTGTCCGAGGAAAAAAAGGACAAATACCGTC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droBip1 | scf7180000396554:1279987-1280030 - | AG-CCTGGTGCCGTTTATCAGAGGAAAAGAAAGATATATACCGCC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droFic1 | scf7180000453172:71145-71188 + | AG-CCTGGTGTGCTATGTCCGAGGAGAAAAAGGATCGCTACCGCC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEle1 | scf7180000489965:1210-1251 + | GGTCCTGGAAACACCTGAGCGAGGAGGAAAAGAGTCGCTACC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droRho1 | scf7180000779483:268370-268372 - | AG-C-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droBia1 | scf7180000302261:1730480-1730523 + | GG-CCTGGTCCAGGCTGCCGGAGGAGAAGAAGGATCGCTACCGCC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droTak1 | scf7180000413216:1263-1304 + | GGTCCTGGAAACACCTGAGCGAGGAGGAAAAGAGTCGCTACC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droEug1 | scf7180000409452:1447544-1447613 - | GG-CCTGGTCCAGGCTCCCGGAGGAGAGAAAGGATCATTACCGCCGCATGGTAAATATATTAGAACATATA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droSim2 | 2l:14440241-14440249 - | GG-CATGGGC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droSec2 | scaffold_3:1151671-1151679 - | AG-CATGGGC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droYak3 | 2L:14984572-14984615 - | AG-CCTGGTCCAAGCTACCCGAGGAAAAAAAGGATCGATACCGCC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEre2 | scaffold_4929:6625113-6625154 - | AG-CCTGGGCCAAGCTACCCGAGGAAAAAAAGGATCGCTATCG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 06/08/2015 at 04:39 PM