ID:dpe_166 |
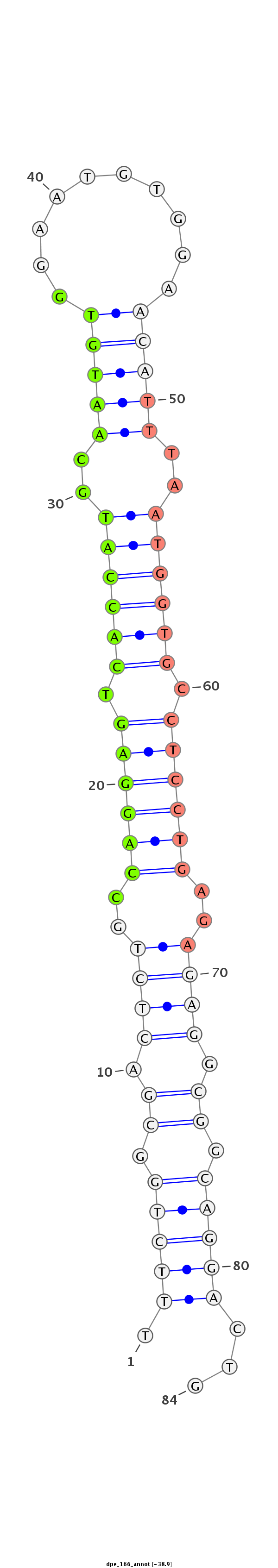
Coordinate:scaffold_26:563931-563985 + |
Confidence:candidate |
Class:Canonical miRNA |
Genomic Locale:intergenic |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -37.9 |
 |
intergenic
No Repeatable elements found
|
TCCTTCCCAACCAAAGAGCCCAGGGAGGAGTTCTGTTTCTGGCGACTCTGCCAGGAGTCACCATGCAATGTGGAATGTGGAACATTTAATGGTGCCTCCTGAGAGAGGCGGCAGGACTGCCATCGGTTGTCTGAGCATTAATCGTCGCGCCTTAT
***********************************.(((((.((.((((..((((((.((((((..(((((..........)))))..)))))).))))))..)))).)).)))))...************************************ |
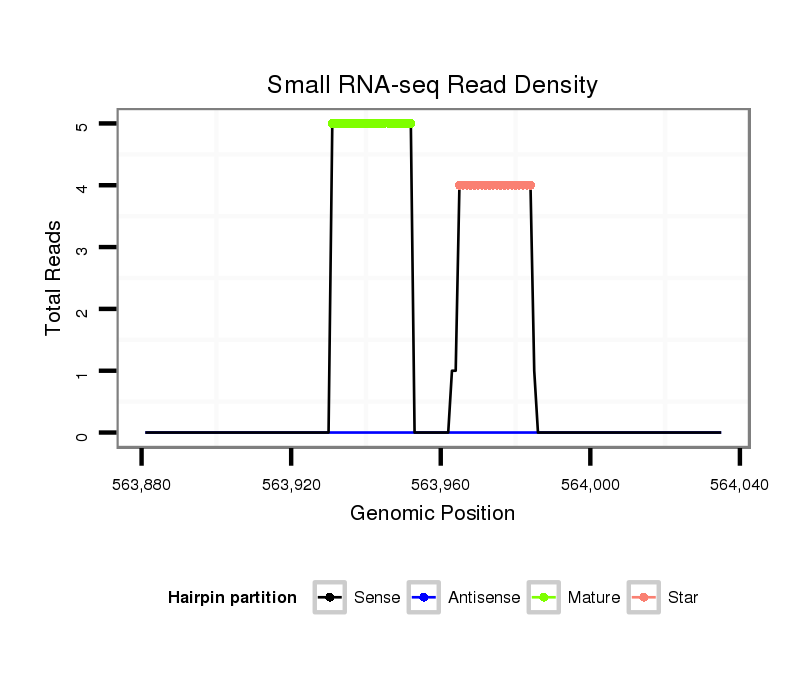
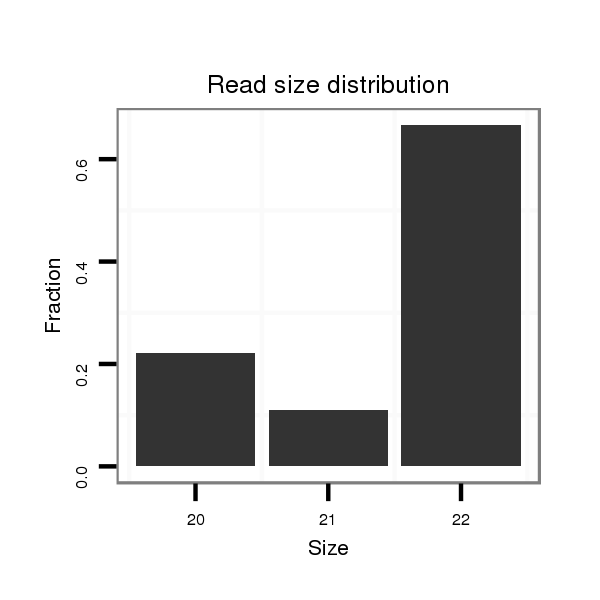
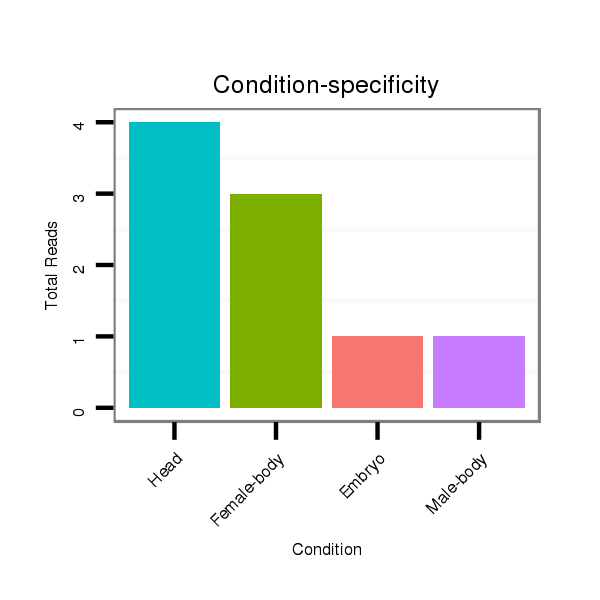
Read size | # Mismatch | Hit Count | Total Norm | Total | V057 head |
M042 female body |
V050 head |
V042 embryo |
V111 male body |
M021 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................CCAGGAGTCACCATGCAATGTG................................................................................... | 22 | 0 | 1 | 5.00 | 5 | 3 | 0 | 0 | 1 | 1 | 0 |
| ....................................................................................TTTAATGGTGCCTCCTGAGA................................................... | 20 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ..CTTCCCACCCAAAGAGCGC...................................................................................................................................... | 19 | 2 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................CATTTAATGGTGCCTCCTGAGA................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................TTTAATGGTGCCTCCTGAGAG.................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................CCAGGAGTCACCATGCAATGTGT.................................................................................. | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................CCAGGAGTCACCATACAAAGAG................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................AGGAGGCAGGACTGCTAACGG............................. | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................TGGGAGGAGTTCTGATACTGG................................................................................................................. | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................ACTCTGGCAGGAGGCACCA............................................................................................ | 19 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..........................................................CACCTCGCAATGTGGAATC.............................................................................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
|
AGGAAGGGTTGGTTTCTCGGGTCCCTCCTCAAGACAAAGACCGCTGAGACGGTCCTCAGTGGTACGTTACACCTTACACCTTGTAAATTACCACGGAGGACTCTCTCCGCCGTCCTGACGGTAGCCAACAGACTCGTAATTAGCAGCGCGGAATA
************************************.(((((.((.((((..((((((.((((((..(((((..........)))))..)))))).))))))..)))).)).)))))...*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V050 head |
M042 female body |
V057 head |
V042 embryo |
M021 embryo |
V111 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................ACCACGGAGGACACTCCCCAC............................................. | 21 | 3 | 6 | 5.50 | 33 | 23 | 10 | 0 | 0 | 0 | 0 |
| .........................................................................................ACCACGGAGGACACTCCCC............................................... | 19 | 2 | 5 | 5.40 | 27 | 14 | 11 | 1 | 0 | 1 | 0 |
| .......................................ACCGCTAATACGGTCCTCAGT............................................................................................... | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................ACCACGGAGGACACTCTC................................................ | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................ACCGCTCATACGGTCCTCAG................................................................................................ | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................ACCACGGAGGACACTCCCCGC............................................. | 21 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................ACCACGGAGGACACTCCCCC.............................................. | 20 | 3 | 13 | 0.77 | 10 | 0 | 10 | 0 | 0 | 0 | 0 |
| ......................................................................................TTTACCACGGAGGACACTC.................................................. | 19 | 2 | 3 | 0.67 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| .........................................................................................ACCACGGAGGACACTCCCCCC............................................. | 21 | 3 | 5 | 0.60 | 3 | 1 | 2 | 0 | 0 | 0 | 0 |
| .......................................ACCGCTACGATGGTCCTCAGT............................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................ACCACGGAGGACTCTCCCAAC............................................. | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................ACCGCTGGGACGGTCCTCAGC............................................................................................... | 21 | 2 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .......................................ACCGCTAGGACGGTCCTCAGT............................................................................................... | 21 | 2 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................ACCACGGAGGACACTCCCCA.............................................. | 20 | 3 | 17 | 0.41 | 7 | 0 | 6 | 1 | 0 | 0 | 0 |
| .........................................................................................ACCACGGAGGACACTCCCCGCT............................................ | 22 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................ACCGCTAAAACGGTCCTC.................................................................................................. | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................ACCGCTAAGACGGTCCTCAGC............................................................................................... | 21 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................ACCGCTGGGACGGGCCTCAGC............................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................ACCGCAGGGACGGTCCTCAGC............................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................ACCGCTAAGACGGTCCTCGGC............................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................CACCGCTGGGACGGTCCTCAGC............................................................................................... | 22 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................ACCGCTAGGACGGTCCTCTGT............................................................................................... | 21 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................CCACGGAGGACACTCCCC............................................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................ACCGATAGGACGGTCCTCAGT............................................................................................... | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................ACCGCTCTGACGGTCCTCTG................................................................................................ | 20 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................ACCGCTGGGACGGTCCTCGGC............................................................................................... | 21 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................ACCCCTGGGACGGTCCTCGG................................................................................................ | 20 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................TTTTCCACGGAGGACACTC.................................................. | 19 | 3 | 17 | 0.12 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| .......................................ACCGCTTGGACGGTCCTCGG................................................................................................ | 20 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................TGTCAACTACCACGGAGGAG...................................................... | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................TCGGGACCCTCTTGAAGAC........................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......................................ACCGCTCGGACTGTCCTCAG................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................ACCGCTGGCACGGTCCTCAG................................................................................................ | 20 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droPer2 | scaffold_26:563881-564035 + | dpe_166 | candidate | TCCTTCCCAACCAAAGAGCCCAGGGAGGAGTTCTGTTTCTGGCGACTCTGCCAGGAGTCACCATGCAATGTGGAA---TGTGGA---------------------------------------------------------------------------ACATTTAATGGTGCCTCCTGAGAGAGGCGGCAGGACTGCCATCGGTTGTCTGAGCATTAATCGTCGCGCCTTAT |
| dp5 | XL_group1a:108363-108595 - | TCCTTCCCAACCAAAGAGCCCAGGGAGGAGTTCTGTTTCTGGCGACTCTGCCAGAAGTCACCATGCAATGTGGAACATTGTGCAAGCCAAGCTATCAAGTTTTACATCAGTTAACATTTAAATACAATAATTTATCTGACCTGGCTTGCCCAAGGTTCCACATTTAATGGTGCCTCCTGAGAGAGGCGGCAGGACTGCCATCGGTTGTCTGAGCATTCATCGTCGCGCCTTAA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
Generated: 10/20/2015 at 06:52 PM