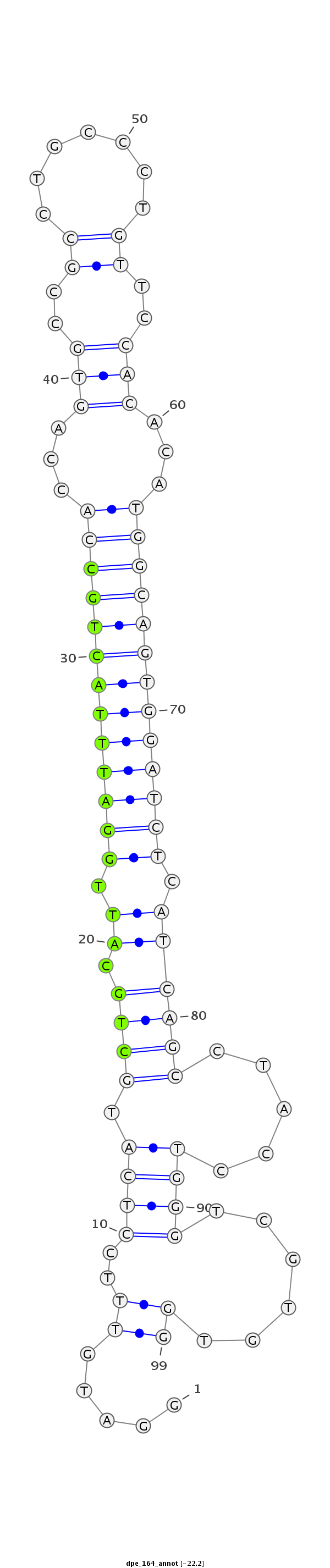
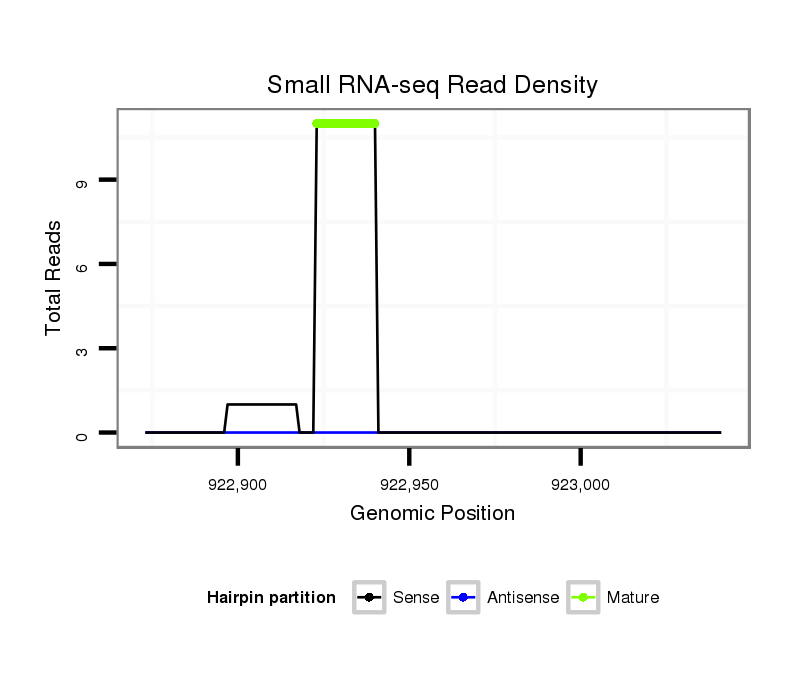
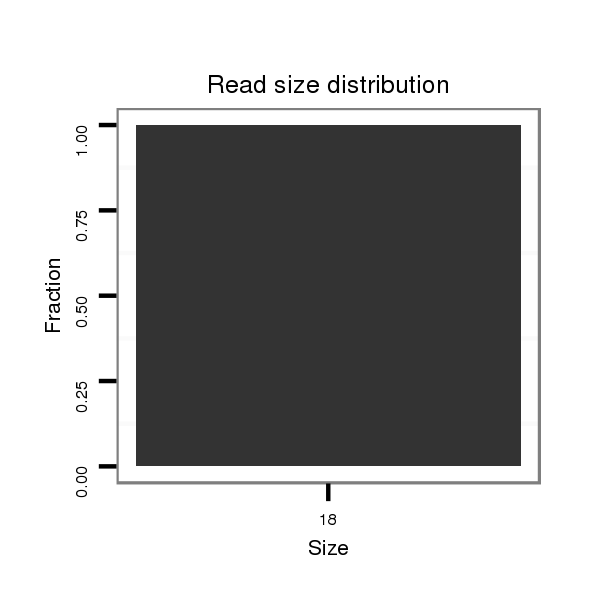
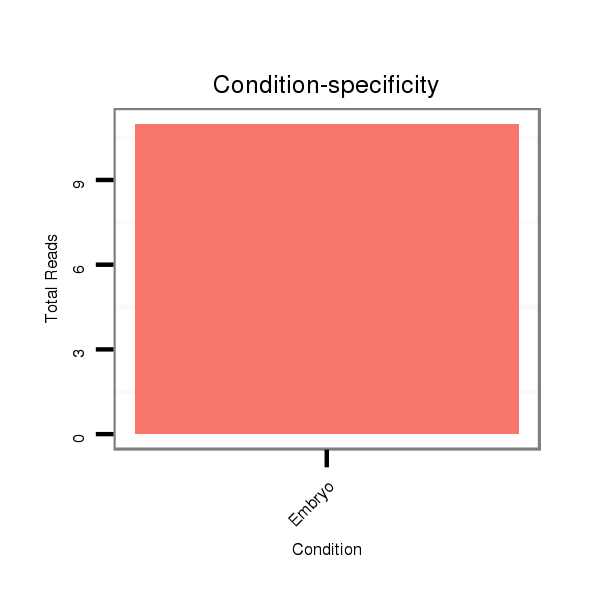
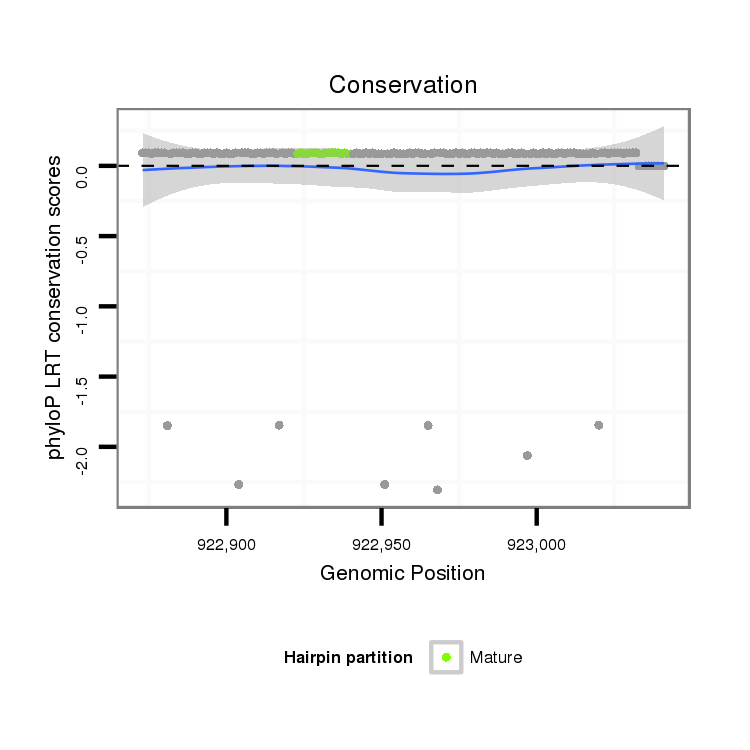
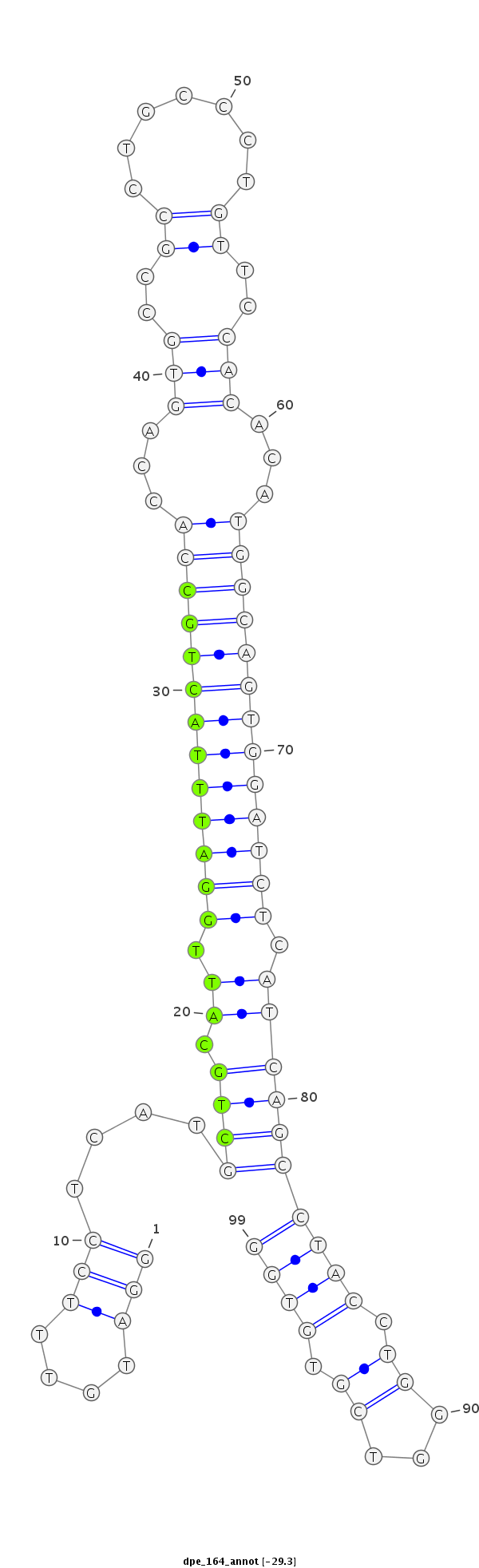
ID:dpe_164 |
Coordinate:scaffold_17:922923-922991 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
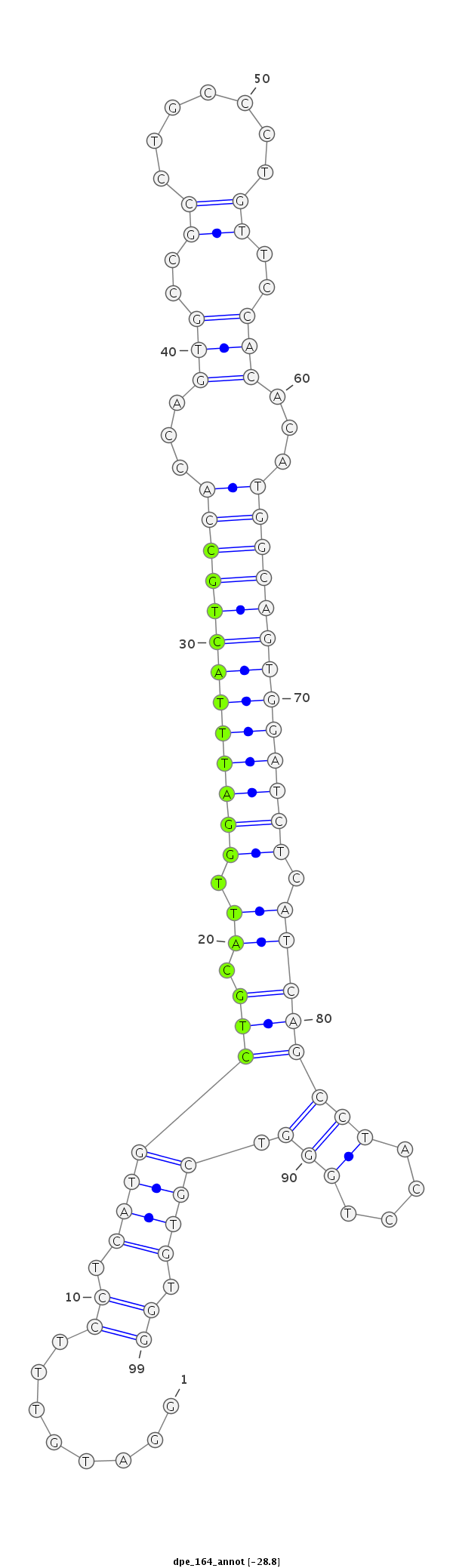
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -29.3 | -28.9 | -28.9 | -28.8 |
 |
 |
 |
 |
intergenic
No Repeatable elements found
| mature | star |
|
TAAGTAAAAGTAAACGATGGAAACTGCACGTGAAAGGATGTTTCCTCATGCTGCATTGGATTTACTGCCACCAGTGCCGCCTGCCCTGTTCCACACATGGCAGTGGATCTCATCAGCCTACCTGGGTCGTGTGGTACTTACCCGCTGCTTCAATTCAAATTCCGTTCTC
***********************************.....((..((((.((((.((.(((((((((((((...(((..((.......))..)))...))))))))))))).)))))).....))))......))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M021 embryo |
V050 head |
V111 male body |
V057 head |
|---|---|---|---|---|---|---|---|---|---|
| ..................................................CTGCATTGGATTTACTGC..................................................................................................... | 18 | 0 | 2 | 11.00 | 22 | 22 | 0 | 0 | 0 |
| ..........................................................................................CCACACATGGCTGCGGGTCTCA......................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ........................TGCACGTGAAAGGATGTTTCC............................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ..................................................CTGCATTGGATTCACTGC..................................................................................................... | 18 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................TGGCAGGGGATCTGATAAGCC................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ..................................................CTGCATTGGATTGACTGC..................................................................................................... | 18 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................CACAGGACAGTGGATCTC.......................................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 |
|
ATTCATTTTCATTTGCTACCTTTGACGTGCACTTTCCTACAAAGGAGTACGACGTAACCTAAATGACGGTGGTCACGGCGGACGGGACAAGGTGTGTACCGTCACCTAGAGTAGTCGGATGGACCCAGCACACCATGAATGGGCGACGAAGTTAAGTTTAAGGCAAGAG
***********************************.....((..((((.((((.((.(((((((((((((...(((..((.......))..)))...))))))))))))).)))))).....))))......))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M042 female body |
|---|---|---|---|---|---|---|
| ..........................................................................................................................ACCCAGCCCGCCAGGAATGG........................... | 20 | 3 | 5 | 0.20 | 1 | 1 |
| ......................................................................................................................................ATGGTTGGGCGACGAAGTT................ | 19 | 2 | 8 | 0.13 | 1 | 1 |
| .....................................................................................................................................AATGGTTGGGCGACGAAGTT................ | 20 | 3 | 10 | 0.10 | 1 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droPer2 | scaffold_17:922873-923041 + | dpe_164 | TAAGTAAAAGTAAACGATGGAAACTGCACGTGAAAGGATGTTTCCTCATGCTGCATTGGATTTACTGCCACCAGTGCCGCCTGCCCTGTTCCACACATGGCAGTGGATCTCATCAGCCTACCTGGGTCGTGT------------------------------GGTACTTACCCGCTGCTTCAATTCAAATTCCGTTCTC |
| dp5 | XL_group1e:3988021-3988210 + | TAAGTAAAGGTAAACGATGGAAACTGCACGTCAAAGGATGTTTCTTCATGCTGCATTGGATTTACTGCCACCAGTGCCCCCTGCCCTGTTCCGCAGATGGCAGTGGATCTCATCAGCCTACCTGTGTCGTGTGGATCGCAGTTATCGAACGGCTTACGGGTTGGTACTTACCCGCTGTTTCAATTCAAAT--------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
|||||||||||||||||||||||||||||||||
| dp5 |
|
Generated: 05/15/2015 at 02:37 PM