
ID:dpe_1536 |
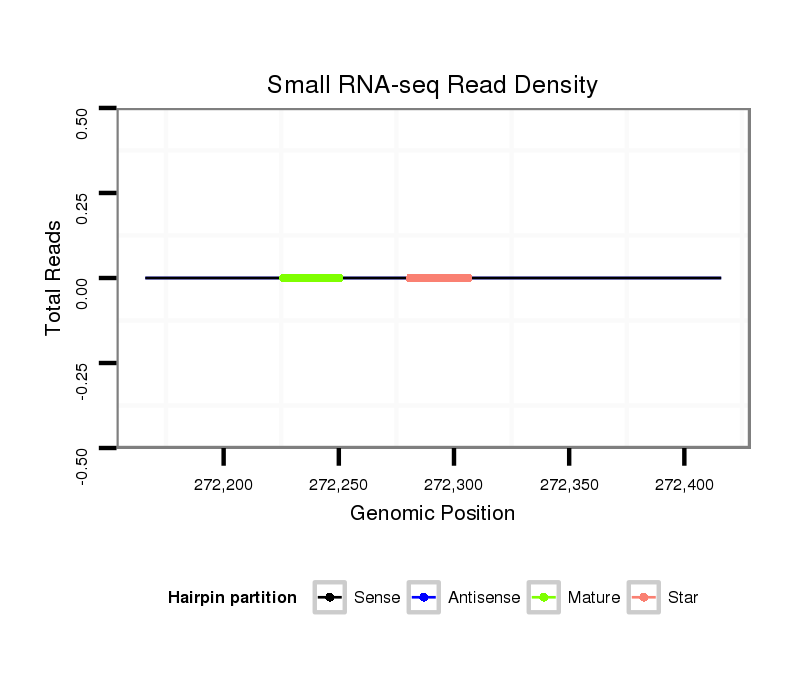
Coordinate:scaffold_38:272216-272366 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
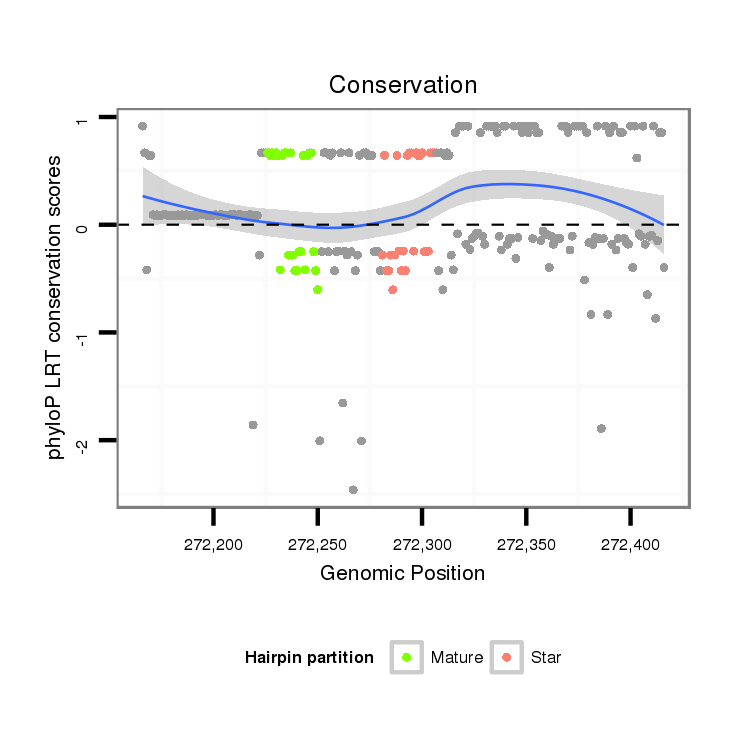
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -22.4 | -21.7 |
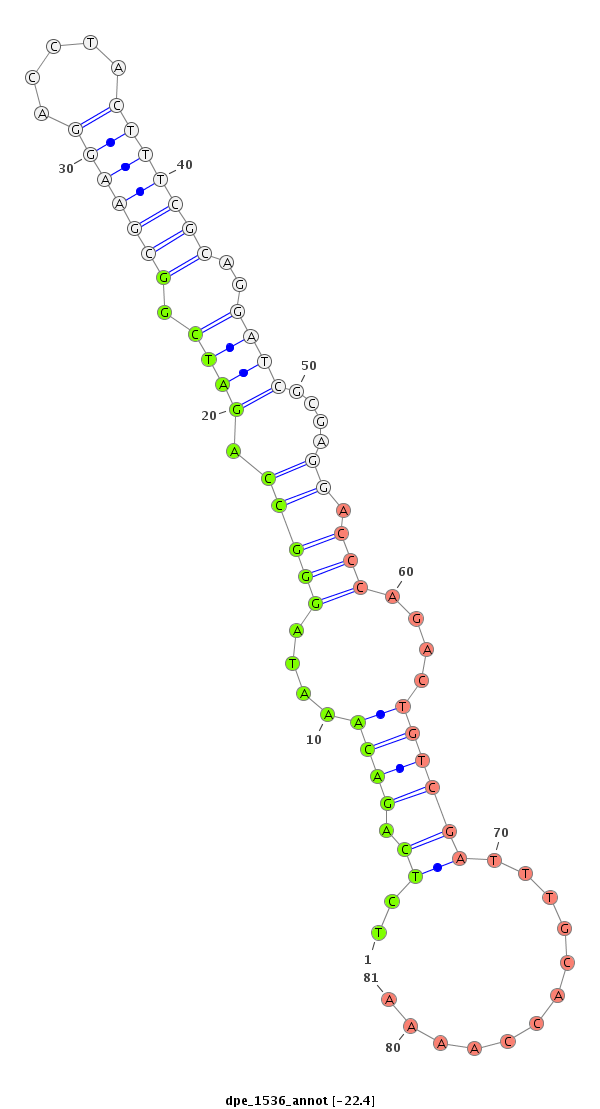 |
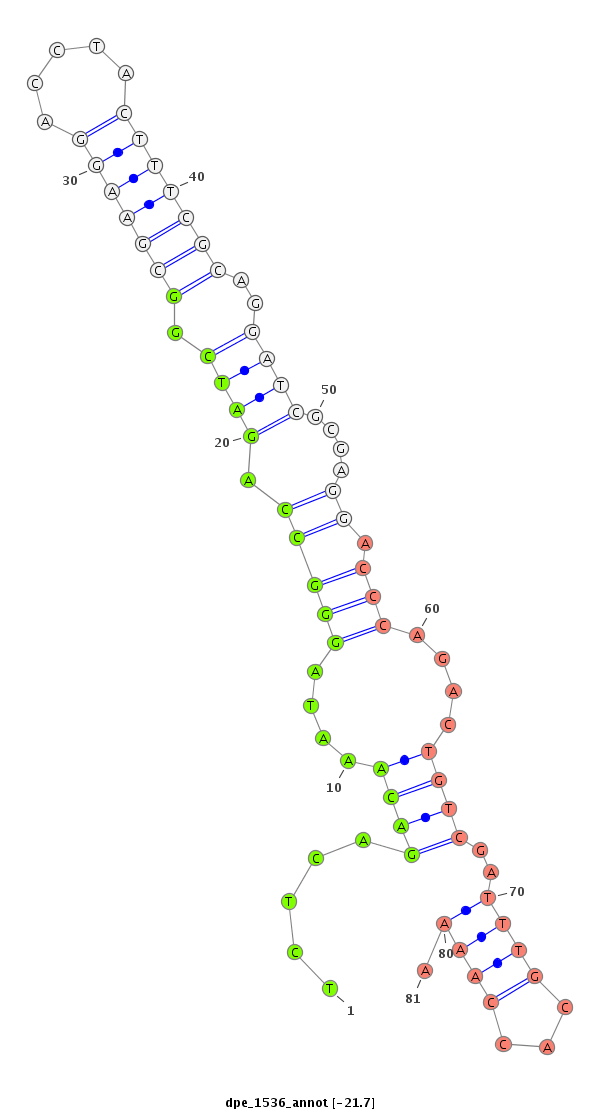 |
exon [dper_GLEANR_13648:2]; CDS [Dper\GL13267-cds]; intron [Dper\GL13267-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TATCCACTCCAAGCGCCTGACTGGCAGATATCGCGACATGCCAATCTTCGGTAGGCTATATCTCAGACAAATAGGGCCAGATCGGCGAAGGACCTACTTTCGCAGGATCGCGAGGACCCAGACTGTCGATTTGCACCAAAATCTCGACTTCGAAAATGAGGCCGATTTTTGGCAAATTTAATGATGTAACCATCATGATTTTTTGCGAAAATCGTGAAAATATGGATGTCCTTCTTTCCGATGGGAGTCGA ************************************************************.....((((....(((((.((((.(((((((.....)))))))..))))....)).)))....))))..............************************************************************************************************************** |
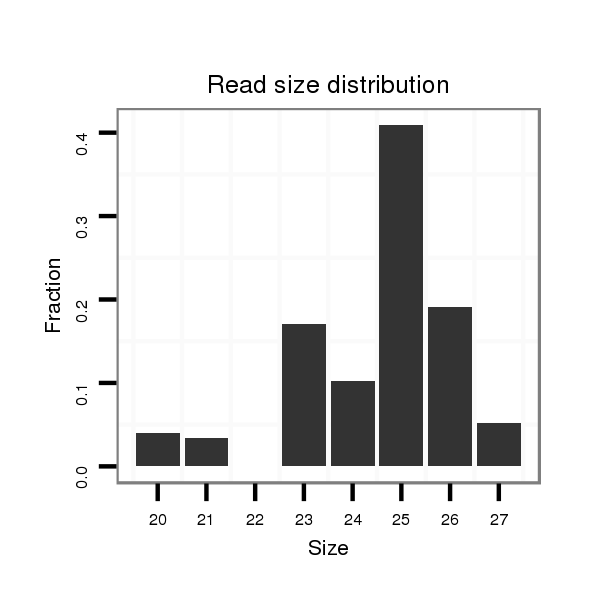
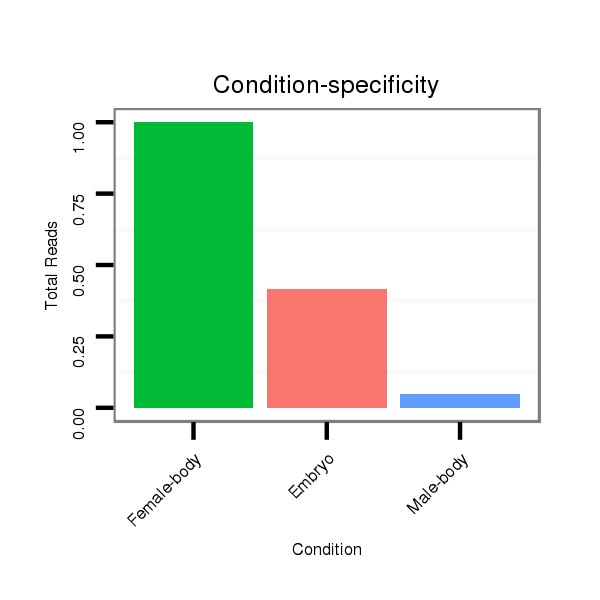
Read size | # Mismatch | Hit Count | Total Norm | Total | M042 female body |
M021 embryo |
V057 head |
V050 head |
V111 male body |
|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................TCTCAGACAAATAGGGCCAGATCGG...................................................................................................................................................................... | 25 | 0 | 20 | 0.50 | 10 | 10 | 0 | 0 | 0 | 0 |
| ...................................................................................................................ACCCAGACTGTCGATTTGCACCAAAA.............................................................................................................. | 26 | 0 | 13 | 0.23 | 3 | 0 | 3 | 0 | 0 | 0 |
| .....................................................................................................................................................TCGAAAATGAGGCCGATTTTTGG............................................................................... | 23 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 |
| .......................................................CTATATCTCAGACAAATAGGGCC............................................................................................................................................................................. | 23 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| .......................................................CTATATCTCAGACAAATAGGGCCA............................................................................................................................................................................ | 24 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................TTTTGCGAAAATCGTGAAAATATGG.......................... | 25 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..............................................................................................TAATTTCGCGGGATCGAGA.......................................................................................................................................... | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................ACCCAGACTGTCGATTTGCACCAAAAT............................................................................................................. | 27 | 0 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................ACCCAGACTGTCGATTTGCACCAAAAAA............................................................................................................ | 28 | 2 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................CCAGACTGTCGATTTGCACC.................................................................................................................. | 20 | 0 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................AAATAGGGCCAGATCGGCGAAGGACC............................................................................................................................................................. | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .............................................CTTCGGTAGGCTATATCTCAG......................................................................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................................................CTATATCTCCGACAAATAGGGCC............................................................................................................................................................................. | 23 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................TCTCGACTTCGAAAATGAGGCCGA...................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................GGCGAAAATCGTGAAAATATGGATG....................... | 25 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................CTCAGACAGATAGGGCCAGATCGG...................................................................................................................................................................... | 24 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................ACTTCGAAAATGAGGCCGATTTTTG................................................................................ | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................GCTATATCTCAGACAAATAGG................................................................................................................................................................................ | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................AATAGGGCCAGATCGGCGAAGGACC............................................................................................................................................................. | 25 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
|
ATAGGTGAGGTTCGCGGACTGACCGTCTATAGCGCTGTACGGTTAGAAGCCATCCGATATAGAGTCTGTTTATCCCGGTCTAGCCGCTTCCTGGATGAAAGCGTCCTAGCGCTCCTGGGTCTGACAGCTAAACGTGGTTTTAGAGCTGAAGCTTTTACTCCGGCTAAAAACCGTTTAAATTACTACATTGGTAGTACTAAAAAACGCTTTTAGCACTTTTATACCTACAGGAAGAAAGGCTACCCTCAGCT
**************************************************************************************************************.....((((....(((((.((((.(((((((.....)))))))..))))....)).)))....))))..............************************************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | M042 female body |
M021 embryo |
V050 head |
V111 male body |
|---|---|---|---|---|---|---|---|---|---|
| ...............................................AGCCATCCGATATAGAGTCTGTT..................................................................................................................................................................................... | 23 | 0 | 18 | 0.22 | 4 | 4 | 0 | 0 | 0 |
| .....................................................................................................................GGACTGATAGCTAAACGTG................................................................................................................... | 19 | 2 | 20 | 0.20 | 4 | 0 | 4 | 0 | 0 |
| .............................................GAAGCCATCCGATATAGAGTCTGTT..................................................................................................................................................................................... | 25 | 0 | 18 | 0.17 | 3 | 3 | 0 | 0 | 0 |
| ............CACGGACTGACCGTCTTTTGC.......................................................................................................................................................................................................................... | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 |
| ..............................................................................................ATTAAAGCGCCCTAGCTCT.......................................................................................................................................... | 19 | 3 | 13 | 0.15 | 2 | 0 | 0 | 0 | 2 |
| .TAGGTGAGGTGCGCAGTCTG...................................................................................................................................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 |
| ................................................GCCATCCGATATAGAGTCTGTT..................................................................................................................................................................................... | 22 | 0 | 18 | 0.11 | 2 | 2 | 0 | 0 | 0 |
| ..............................................AAGCCATCCGATATAGAGTCTGTT..................................................................................................................................................................................... | 24 | 0 | 18 | 0.11 | 2 | 2 | 0 | 0 | 0 |
| ...................TGCCCTTCCATAGCGCTG...................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 |
| .........................................GTTAGAAGCCATCCGATATAGAGT.......................................................................................................................................................................................... | 24 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| ...........................................TAGAAGCCATCCGATATAGAGTCTGTT..................................................................................................................................................................................... | 27 | 0 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| .....................................TACGGTTAGAAGCCATCCGATATAGAGT.......................................................................................................................................................................................... | 28 | 0 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ......................................ACGGTTAGAAGCCATCCGATATAGAGT.......................................................................................................................................................................................... | 27 | 0 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ............................................AGAAGCCATCCGATATAGAGTCTGTT..................................................................................................................................................................................... | 26 | 0 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| .......................................CGGTTAGAAGCCATCCGATATAGAGT.......................................................................................................................................................................................... | 26 | 0 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ..............................................AAGCCATCCGATATAGAGTCTGT...................................................................................................................................................................................... | 23 | 0 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| .............................................AAAGCCATCCGATATAGAGTCTGT...................................................................................................................................................................................... | 24 | 1 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| .......................................CGGTTAGAAGCCATCCGATATAGAG........................................................................................................................................................................................... | 25 | 0 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................................................CTGACAGCTAAACGTGGTTTT.............................................................................................................. | 21 | 0 | 19 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ........................................TGTTAGAAGCCATCCGATA................................................................................................................................................................................................ | 19 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ........................................GGTTAGAAGCCATCCGATATAGAGT.......................................................................................................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droPer2 | scaffold_38:272166-272416 + | dpe_1536 | TATCCACTCCAAGCGCCTGACTGGCAGATATCGCGACATGCCAATCTTCGGTAGGCTATATCTCAGACAAATAGGGCCAGATCGGCGAAGGACCTACTTTCGCAGGATCGCGAGGACCCAGACTGTCGATTTGCACCAAAATCTCGACTTCGAAAATGAGGCCGATTTTTGGCAAATTTAATGATGTAACCATCAT-GATTTTTTGCGAAAATCG--TGAAAATATGGATGTCCTTC-----TTTCCGATGGGAGTCGA |
| dp5 | XR_group8:5414350-5414600 - | dps_3331 | TATCCACTCCAAGCGCCTGACTGGCAGATATCGCGACATGCCAATCTTCGGTAAGCTATATCTCAGACAAATAGGGCCAGATCGGTGAAGGACCTATTTTCCCAGAATCGCGAGGACCCAGACTGTCGATTTGCACCAAAATCTCGACTTCGAAAATGAGGCCGATTTTTGGCAAATTTAATGATGTAACCATCAT-GATTTTTTGCGAAAATCG--TGAAAAAATGGATGTCCTTC-----TTTCCGATGGCAGTCGA |
| droAna3 | scaffold_13340:16404276-16404476 - | TAGCC---------------------------------------------------AATATCTCAGCCAATTTTTATCCGATTTCAAAAAGAATCACCATTTATGTATCGCAGATTCAATCTCCTCAGATCTGCATTGAAATATGGACAGCAAAAATTTGATCGATTTTTGGCAAAAATCGTGATGTTACCCCTTT-GAAAATTTGCGAAAATCGGCTCAGATTTTCGATGACCATA-----TTT-TGATTGGAAGCGA | |
| droKik1 | scf7180000302686:1405839-1405946 + | T-----------------------------------------------------------------------------------------------------------------------------------------------------CAAAATTTAAGCCATTTTTTGAAAATTTTAATGATGTAACCCCTTGCATTTTTTTGAAAAAATGG--CTTAAAAATTGTTTTCCTCCGGACATGTCTAAAAAGACGCGC |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
Generated: 06/08/2015 at 04:23 PM