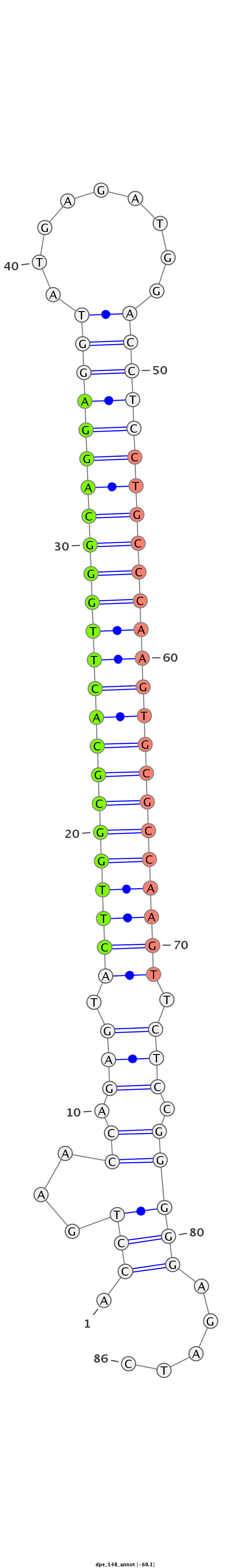
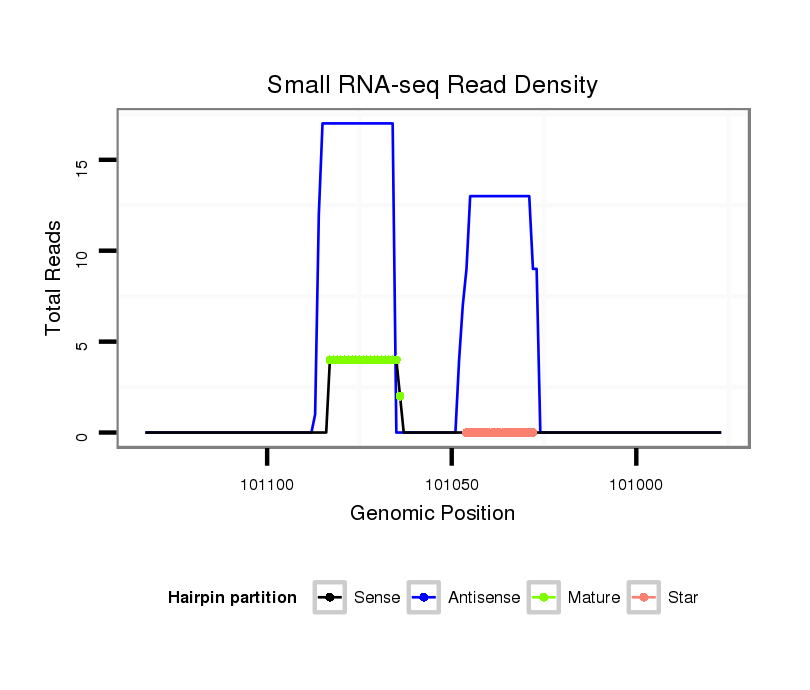
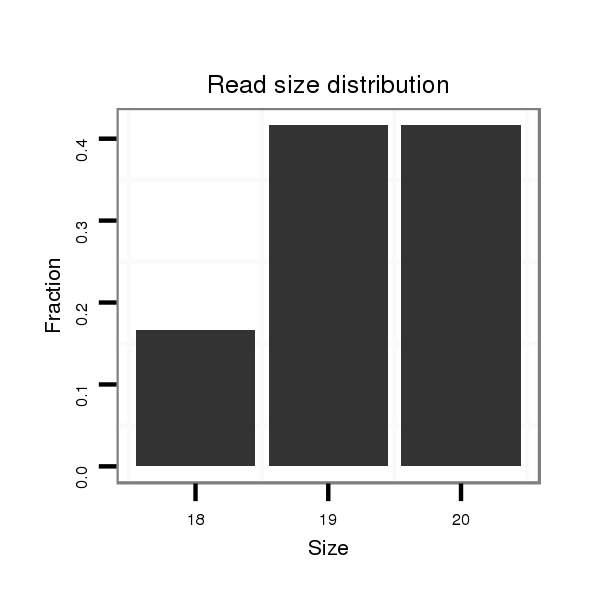
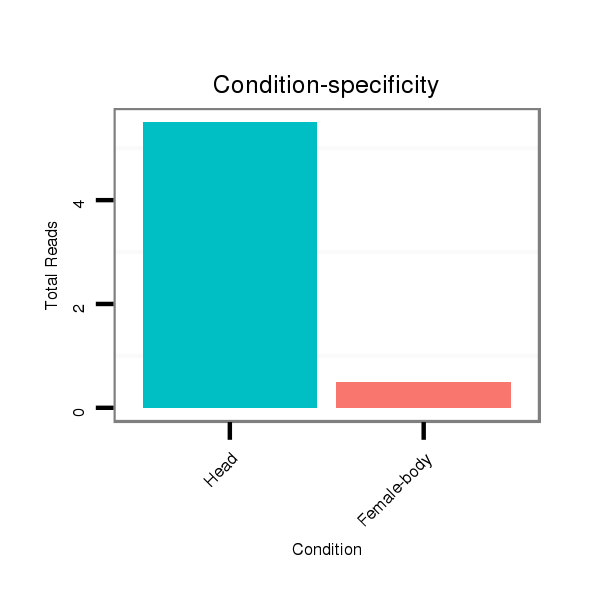
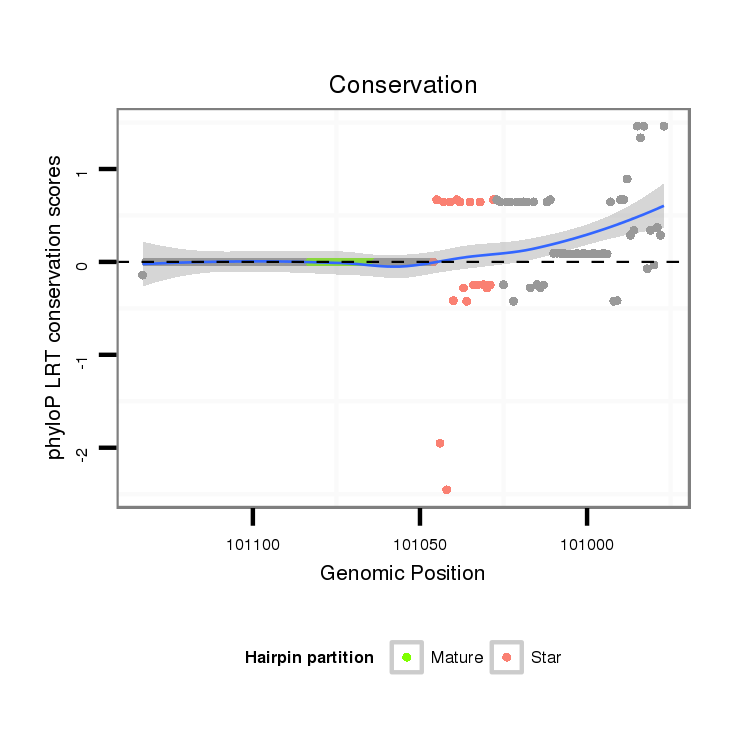
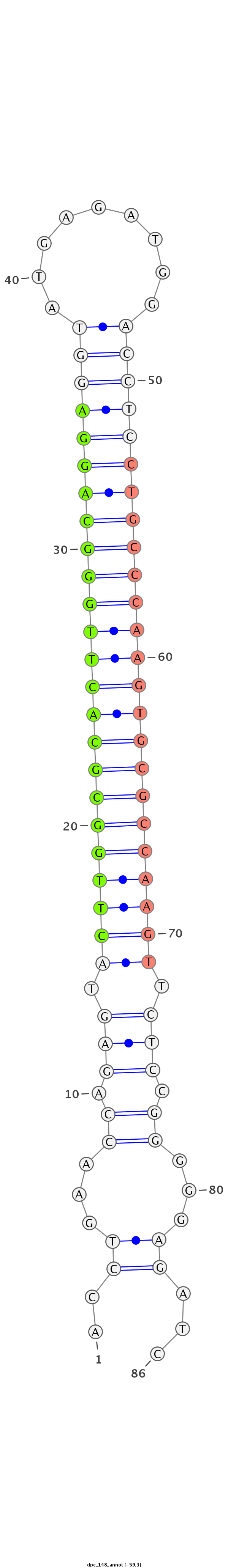
ID:dpe_148 |
Coordinate:scaffold_26:101027-101083 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -59.3 |
 |
intergenic
No Repeatable elements found
| mature | star |
|
TGTAAATCCCTCAGTGTAAAGAGAATTCTGGCCTGACCTGAACCAGAGTACTTGGCGCACTTGGGCAGGAGGTATGAGATGGACCTCCTGCCCAAGTGCGCCAAGTTCTCCGGGGGAGATCGTTAATGGGTGTGCAGGAGCGATTCGATGTTTTCTT
***********************************.(((...((.(((.((((((((((((((((((((((((.........)))))))))))))))))))))))).))).))))).....************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | V050 head |
V057 head |
M042 female body |
|---|---|---|---|---|---|---|---|---|
| ..................................................CTTGGCGCACTTGGGCAGGA....................................................................................... | 20 | 0 | 2 | 2.50 | 5 | 4 | 0 | 1 |
| ..................................................CTTGGCGCACTTGGGCAGG........................................................................................ | 19 | 0 | 2 | 2.00 | 4 | 2 | 2 | 0 |
| .......................................................................................CTGCCCAAGTGCGCCAAGT................................................... | 19 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 |
| ...................................................TTGGCTCACTTGGGCAGGA....................................................................................... | 19 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 |
| ..................................................CTTGGCGCGCTTGGGCAGGA....................................................................................... | 20 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 |
| ...................................................TTGGCGCACTTGGGCAGG........................................................................................ | 18 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 |
| .................................................ACTTGGCGCACTTGGGCAGG........................................................................................ | 20 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 |
| .................................................ACTTGGCGTACTTGGGCAGGAG...................................................................................... | 22 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 |
| ....................................................TGGCGCACTTGGGCAGGA....................................................................................... | 18 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| ..................................................CTTGGCGCACTTGAGCAGGA....................................................................................... | 20 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| ..................................................CGGGGCGCACTTGGGCAGGT....................................................................................... | 20 | 3 | 14 | 0.07 | 1 | 0 | 1 | 0 |
|
ACATTTAGGGAGTCACATTTCTCTTAAGACCGGACTGGACTTGGTCTCATGAACCGCGTGAACCCGTCCTCCATACTCTACCTGGAGGACGGGTTCACGCGGTTCAAGAGGCCCCCTCTAGCAATTACCCACACGTCCTCGCTAAGCTACAAAAGAA
************************************.(((...((.(((.((((((((((((((((((((((((.........)))))))))))))))))))))))).))).))))).....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V050 head |
V057 head |
V042 embryo |
V111 male body |
M042 female body |
|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................CATGAACCGCGTGAACCCGTC......................................................................................... | 21 | 0 | 1 | 11.00 | 11 | 5 | 5 | 1 | 0 | 0 |
| ................................................ATGAACCGCGTGAACCCGTC......................................................................................... | 20 | 0 | 1 | 5.00 | 5 | 3 | 0 | 0 | 2 | 0 |
| ........................................................................................ACGGGTTCACGCGGTTCAA.................................................. | 19 | 0 | 1 | 4.00 | 4 | 2 | 2 | 0 | 0 | 0 |
| .....................................................................................AGGACGGGTTCACGCGGTTC.................................................... | 20 | 0 | 2 | 2.50 | 5 | 4 | 0 | 0 | 0 | 1 |
| .......................................................................................GACGGGTTCACGCGGTTCAA.................................................. | 20 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 |
| .....................................................................................AGGACGGGTTCACGCGGTTCAA.................................................. | 22 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 |
| ......................................................................................GGACGGGTTCACGCGGTTC.................................................... | 19 | 0 | 2 | 2.00 | 4 | 2 | 2 | 0 | 0 | 0 |
| ......................................................................................GGACGGGTTCACGCGGTTCAA.................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................TCATGAACCGCGTGAACCCGTC......................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................TGACGGGTTCACGCGGTGCAA.................................................. | 21 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................TCATGAACCGGGTGAACACGTC......................................................................................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............................................TATGAACCGCGTGAACCCGTC......................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................ACGGGTACACGCGGTTCAA.................................................. | 19 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................ATGCACCGCGTGAACCCGTC......................................................................................... | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................TGAACCGCGTGAACCCGTC......................................................................................... | 19 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................GAGGACGGGTTCATGCGGTTCA................................................... | 22 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................AGGACGGGTTCGCGCGGTTC.................................................... | 20 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................AGGACGGGTTCACGCGGT...................................................... | 18 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................GGACGGGTTCACGCGGTT..................................................... | 18 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................AGGACGAGTTCACGCGGTTC.................................................... | 20 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................AGGACGGGTTCACTCGGTT..................................................... | 19 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 |
| ......................................................................................GGACGGGTTCACGCGGTTCA................................................... | 20 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................TGGACGGGTTCACGCGGGGC.................................................... | 20 | 3 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................GGTTCACGCAGTTCGAGA................................................ | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................CGCGCTTCAATCGGCCCC.......................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droPer2 | scaffold_26:100977-101133 - | dpe_148 | TGTAAATCCCTCAGTGTAAAGAGAATTCTGGCCTGACCTGAACCAGAGTACTTGGCGCACTTGGGCAGGAGGTATGAGATGGACCTCCTG-CCCAAGTGCGCCAAGTTCTCCGGGGGAGATCGTTAATGGGTGTGCAGGAGCGATTCGATGTTTTC---TT |
| dp5 | XL_group1a:494883-494952 - | C---------------------------------------------------------------------------------------TT-CGCAAGTGCGCCAAGTTCTCCGGGGGAGATCGTTAATGGGTGTGCAGGAGCGATTCGATGTTTTC---TT | |
| droAna3 | scaffold_12929:1713906-1713916 + | ---------------------------------------------------------------------------------------------------------------------------------------------------GATGTCTTT---GT | |
| droBip1 | scf7180000396543:179203-179259 + | C---------------------------------------------------------------------------------------TTGCTCCAGATCATCGTATTCCCCTGGGGTGGATGT-----------------CTCTTCTGTGTTCTTTGTGT | |
| droEle1 | scf7180000491011:1091594-1091605 - | C--------------------------------------------------------------------------------------------------------------------------------------------------GATGTCTCT---TT | |
| droRho1 | scf7180000779506:879252-879263 - | C--------------------------------------------------------------------------------------------------------------------------------------------------GATGTCTTT---TT | |
| droBia1 | scf7180000301760:1057026-1057037 - | C--------------------------------------------------------------------------------------------------------------------------------------------------GATGTCTTT---TT | |
| droTak1 | scf7180000415179:44314-44327 - | T-------------------------------------------------------------------------------------------------------------------------------------------------CGATGTCTTT--CTT | |
| droEug1 | scf7180000409277:421552-421563 - | C--------------------------------------------------------------------------------------------------------------------------------------------------GATGTCTCT---TT |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
Generated: 05/15/2015 at 02:37 PM