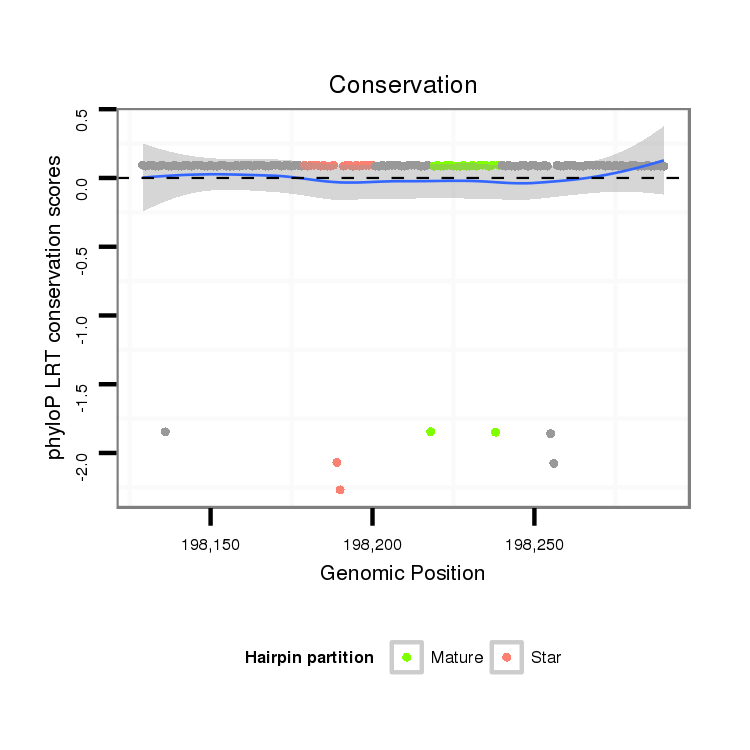
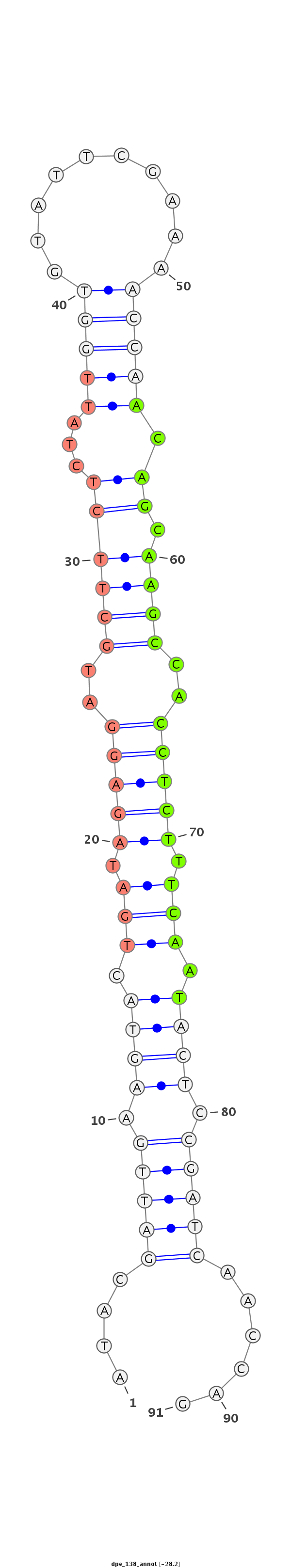
ID:dpe_138 |
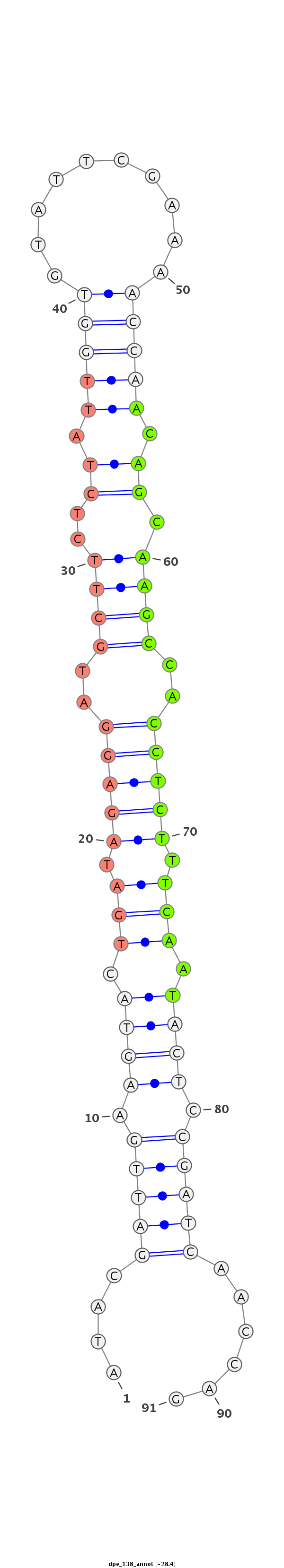
Coordinate:scaffold_13:198179-198240 + |
Confidence:confident |
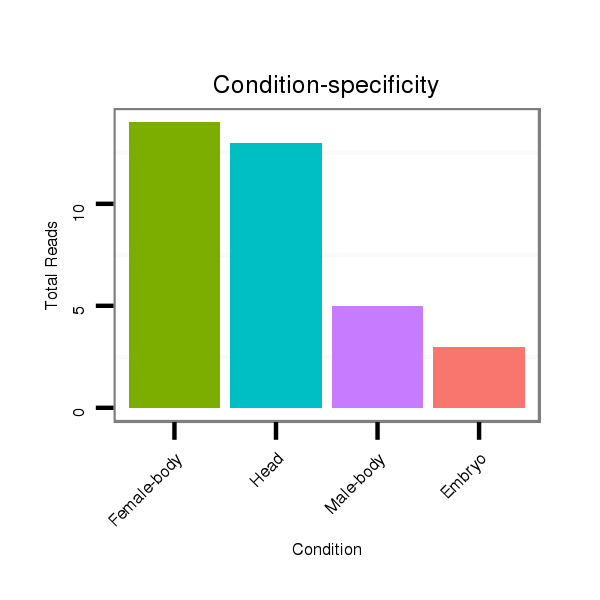
Class:Testes-restricted |
Genomic Locale:intergenic |
[View on UCSC Genome Browser {Cornell Mirror}] |
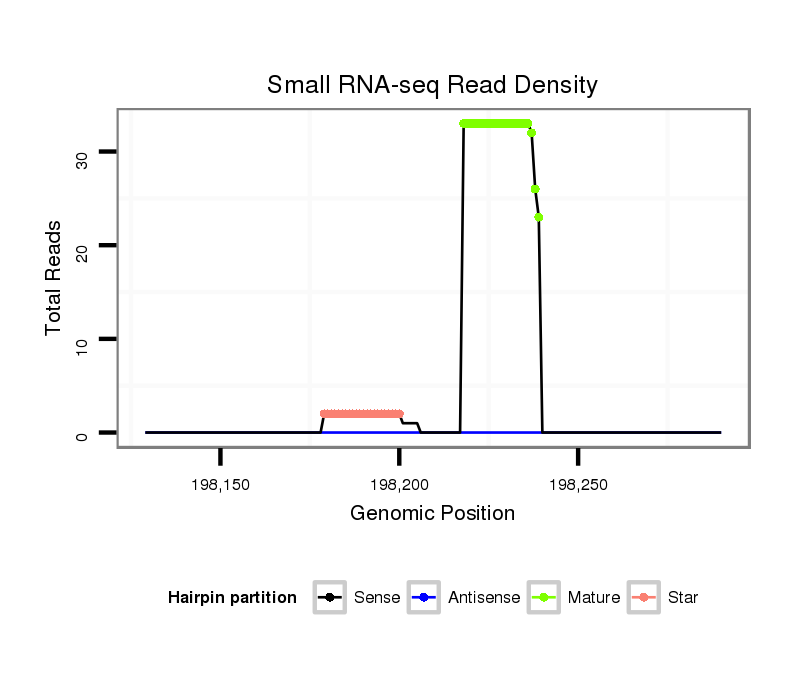
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -28.2 |
 |
intergenic
No Repeatable elements found
|
AGATCGACCGACGATCTATGGAGGATCCCTGTTTCATACGATTGAAGTACTGATAGAGGATGCTTCTCTATTGGTGTATTCGAAAACCAACAGCAAGCCACCTCTTTCAATACTCCGATCAACCAGTCTGGTATCCCAATATTCTGTGACGAACGCTGGAGG
***********************************....(((((.((((.(((.(((((..((((..((.(((((..........))))).)).))))..))))).))).)))).)))))......************************************ |
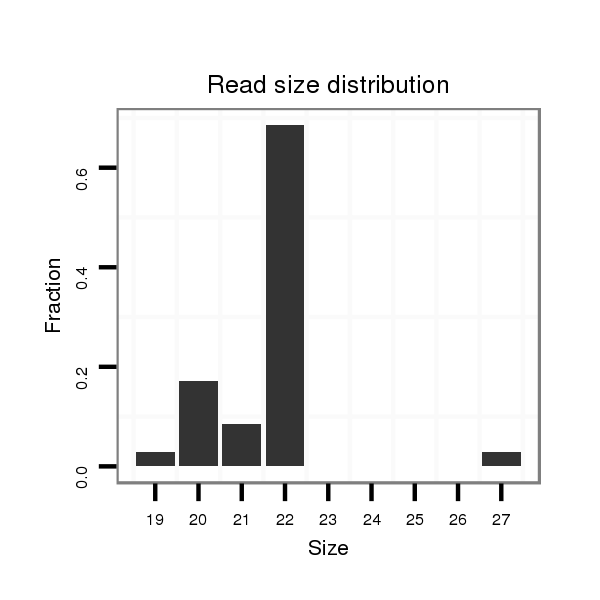
Read size | # Mismatch | Hit Count | Total Norm | Total | V057 head |
M042 female body |
V111 male body |
M021 embryo |
V050 head |
|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................ACAGCAAGCCACCTCTTTCAAT................................................... | 22 | 0 | 1 | 23.00 | 23 | 6 | 11 | 3 | 3 | 0 |
| .........................................................................................ACAGCAAGCCACCTCTTTCA..................................................... | 20 | 0 | 1 | 6.00 | 6 | 3 | 1 | 1 | 0 | 1 |
| .........................................................................................ACAGCAAGCCACCTCTTTCAA.................................................... | 21 | 0 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 | 0 |
| .........................................................................................ACAGCAAGCCACCTCTTTC...................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................ACAGCAGGCCACCTCTTTCAAT................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................ACAGCCAGCCACCTCTTTCAAT................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TGATAGAGGATGCTTCTCTATT.......................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TGATAGAGGATGCTTCTCTCT........................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................TGATAGAGGATGCTTTTCTATTG......................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................ACACCAAGCCACCTCTTCCA..................................................... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TGATAGAGGATGCTTCTCTATTGGTGT..................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................TGATAGAGGATGCTTCTCTATTGA........................................................................................ | 24 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................TGATAGAAGATGGTTCTCTATT.......................................................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TGATAGAGGATGCTTCTCTATTC......................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................ACAGAAAGCCACCTCTTTCAAT................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TGATAGAGGATGCTTCTCTTT........................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................TGATAGAGGATGATTCTCCATTG......................................................................................... | 23 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TGATAGAGGATGCTTCTTTCT........................................................................................... | 21 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................................................GATAGAGGATACTTCTCCAATGGT....................................................................................... | 24 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................ACAAGAAGCCACCTCTTTCACT................................................... | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TGATAGAGGATGCTTCTCCT............................................................................................ | 20 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................TGATAGAGGATGCTTTTTT............................................................................................. | 19 | 2 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 |
|
TCTAGCTGGCTGCTAGATACCTCCTAGGGACAAAGTATGCTAACTTCATGACTATCTCCTACGAAGAGATAACCACATAAGCTTTTGGTTGTCGTTCGGTGGAGAAAGTTATGAGGCTAGTTGGTCAGACCATAGGGTTATAAGACACTGCTTGCGACCTCC
************************************....(((((.((((.(((.(((((..((((..((.(((((..........))))).)).))))..))))).))).)))).)))))......*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V057 head |
|---|---|---|---|---|---|---|
| .........................................................................................................................TGGTCAGACTGTACGGTTATA.................... | 21 | 3 | 3 | 0.33 | 1 | 1 |
| ....................................................................................................GGAGATAGGTACGAGGCTAGT......................................... | 21 | 3 | 3 | 0.33 | 1 | 1 |
| ....................................................................................................GGAGACAGGTACGAGGCTAGT......................................... | 21 | 3 | 3 | 0.33 | 1 | 1 |
| ....................................................................................................GGAGAAAGTCAAGAGGCTAGT......................................... | 21 | 2 | 3 | 0.33 | 1 | 1 |
| ....................................................................................................GGAAAAAGTCACGAGGCTAGT......................................... | 21 | 3 | 7 | 0.14 | 1 | 1 |
| ....................................................................................................GTAGAAAGTCAAGAGGCTAGT......................................... | 21 | 3 | 8 | 0.13 | 1 | 1 |
| ......................................................................................................................AGTTGGTGAGCCCGTAGGG......................... | 19 | 3 | 20 | 0.05 | 1 | 1 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droPer2 | scaffold_13:198129-198290 + | dpe_138 | confident | AGATCGACCGACGATCTATGGAGGATCCCTGTTTCATACGATTGAAGTACTGATAGAGGATGCTTCTCTATTGGTGTATTCGAAAACCAACAGCAAGCCACCTCTTTCAATACTCCGATCAACCAGTCTGGTATCCCAATATTCTGTGACGAACGCTGGAGG |
| dp5 | XL_group1e:8366108-8366269 - | dps_62 | confident | AGATCGATCGACGATCTATGGAGGATCCCTGTTTCATACGATTGAAGTACTGATAGAGGAGCCTTCTCTATTGGTGTATTCGAAAACCATCAGCAAGCCACCTCTTTCAGTACTCCGATCAACCAGCATGGTATCCCAATATTCTGTGACGAACGCTGGAGG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
Generated: 10/20/2015 at 07:37 PM