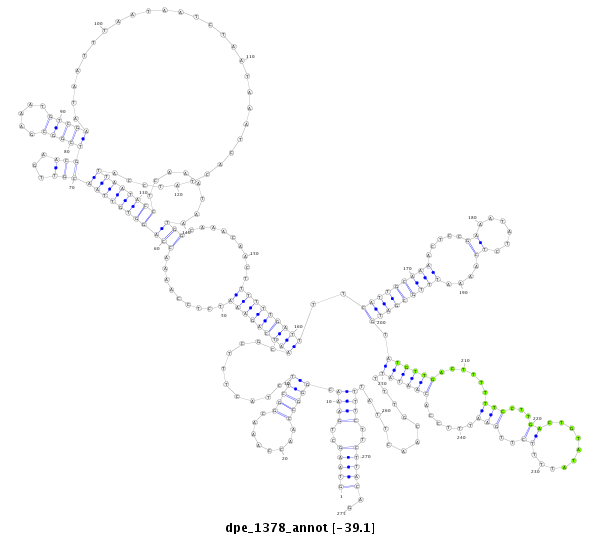
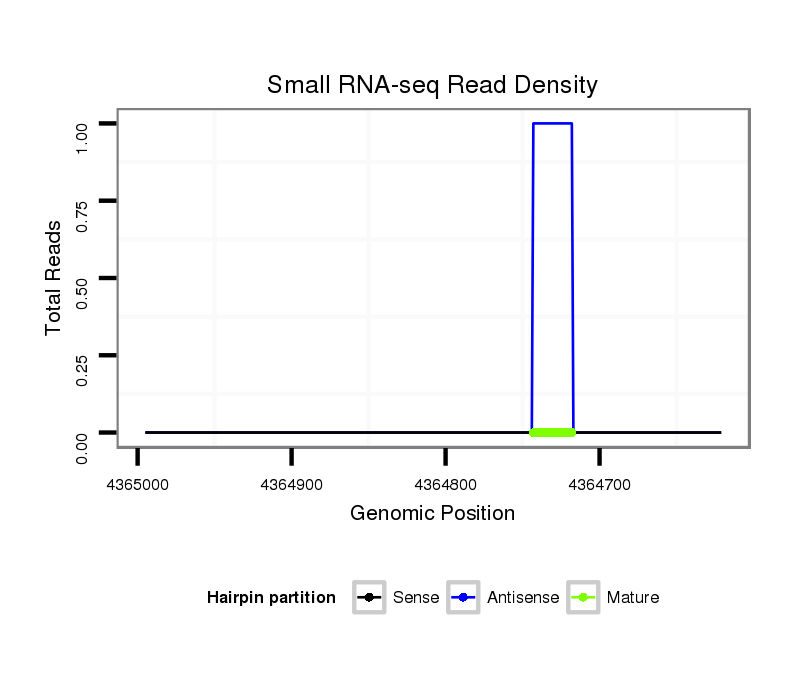
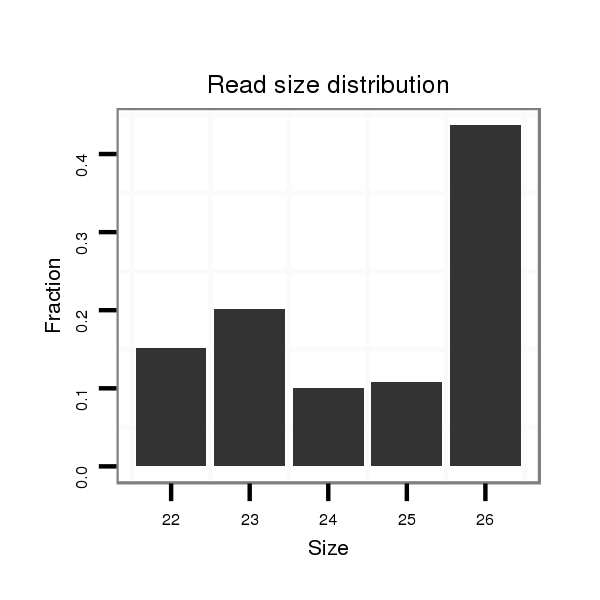
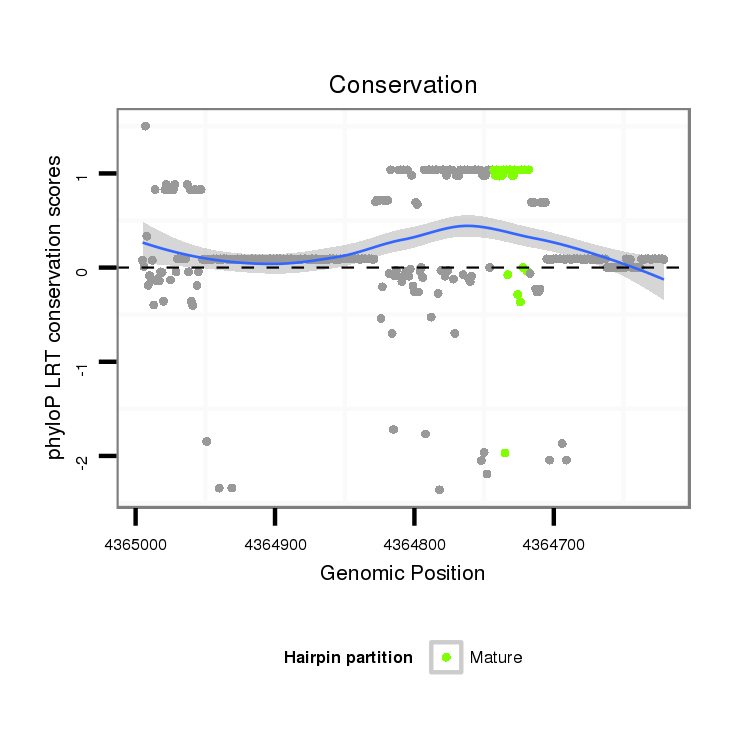
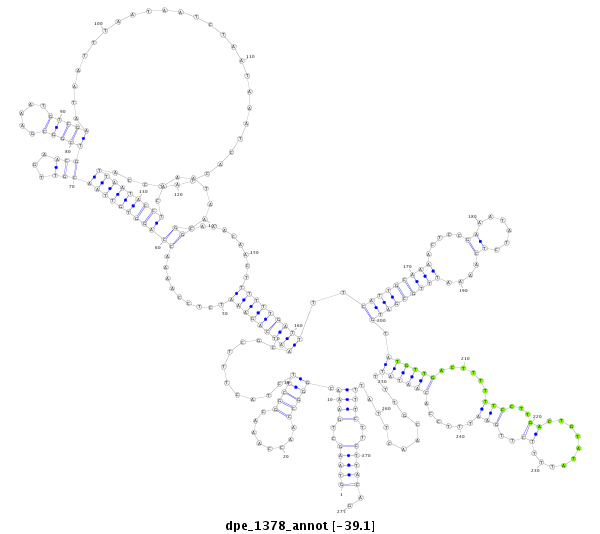
| TAATGATTC---------------------------------------------------------------------------------------------------------------------------------------------------------------------CTATGTATTAATGTTCTAACTATTTGCC-----TTATTTTTTATTTTCGTTTCGAAACCCCGAAATATCGAAAAATTGGTGGTATATGTTGACTATTTTCCTTCACTATAAAT-------------------------------------------------------------------------------------------------------------------------- | Size | Hit Count | Total Norm | Total | M046
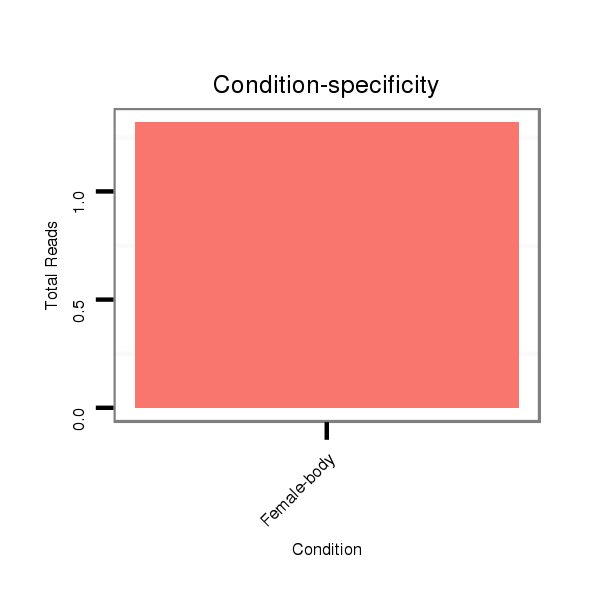
Female-body | V041
Embryo | V049
Head | V056
Head | M060
Embryo | V110
Male-body |
| ................................................................................................................................................................................................................................................................TATATGTTGACTATTTTCCTTCACTA............................................................................................................................... | 26 | 1 | 24.00 | 24 | 23 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................................................................TATATGTTGACTATTTTCCTTCACT................................................................................................................................ | 25 | 1 | 20.00 | 20 | 14 | 0 | 0 | 0 | 6 | 0 |
| ...........................................................................................................................................................................................TTCTAACTATTTGCC-----TTATTTTTTATT.............................................................................................................................................................................................. | 27 | 1 | 6.00 | 6 | 6 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................TATATGTTGACTATTTTCCTTCACTATA............................................................................................................................. | 28 | 1 | 5.00 | 5 | 3 | 0 | 0 | 0 | 2 | 0 |
| ................................................................................................................................................................................................................................................................TATATGTTGACTATTTTCCTTCACC................................................................................................................................ | 25 | 1 | 4.00 | 4 | 0 | 0 | 0 | 0 | 4 | 0 |
| .........................................................................................................................................................................................................................................................TTGGTGGTATATGTTGACTATTTTCC...................................................................................................................................... | 26 | 1 | 4.00 | 4 | 3 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................................................................TATATGTTGACTATTTTCCTTCACTAT.............................................................................................................................. | 27 | 1 | 4.00 | 4 | 2 | 0 | 0 | 0 | 2 | 0 |
| ................................................................................................................................................................................................................................................................TATATGTTGACTATTTTCCTTCAC................................................................................................................................. | 24 | 1 | 3.00 | 3 | 1 | 0 | 0 | 0 | 2 | 0 |
| ................................................................................................................................................................................................................................TTCGAAACCCCGAAATATCGAAAAATTG............................................................................................................................................................. | 28 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................CTAACTATTTGCC-----TTATTTTTTATC.............................................................................................................................................................................................. | 25 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................TCGAAACCCCGAAATATCGAAAAATTGG............................................................................................................................................................ | 28 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................TATTTTCGTTTCGAAACCCCGAAATA........................................................................................................................................................................ | 26 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................................................TTCTAACTATTTGCC-----TTATTTTTTATTT............................................................................................................................................................................................. | 28 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................TATATGTTGACTATTTTCCTTCACTATAA............................................................................................................................ | 29 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................GTATATGTTGACTATTTTCCTTCACTA............................................................................................................................... | 27 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................TCTAACTATTTGCC-----TTATTTTTTA................................................................................................................................................................................................ | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................TCGTTTCGAAACCCCGAAATATCGAA................................................................................................................................................................... | 26 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................CGAAACCCCGAAATATCGAAAAATTGG............................................................................................................................................................ | 27 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................TGTTGACTATTTTCCTTCACTATA............................................................................................................................. | 24 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |