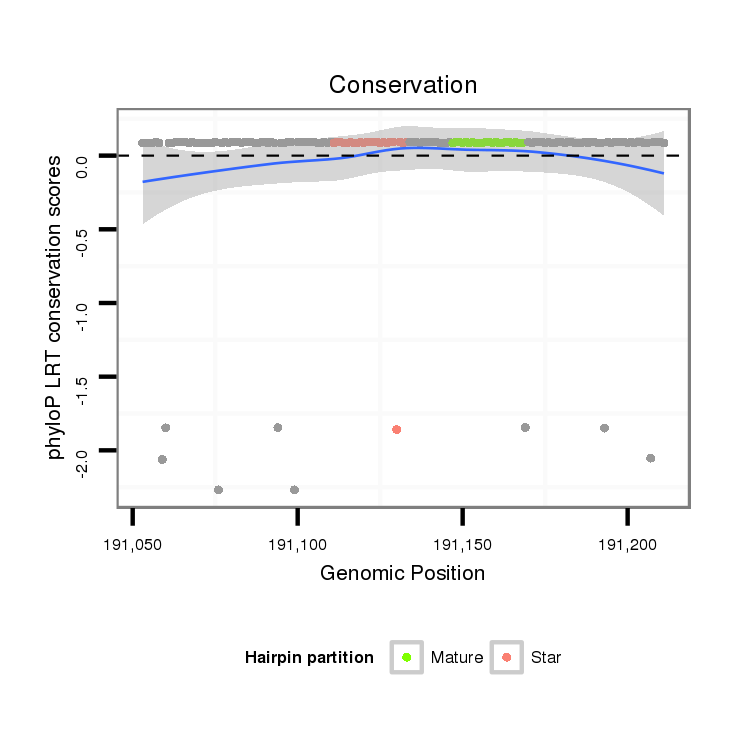
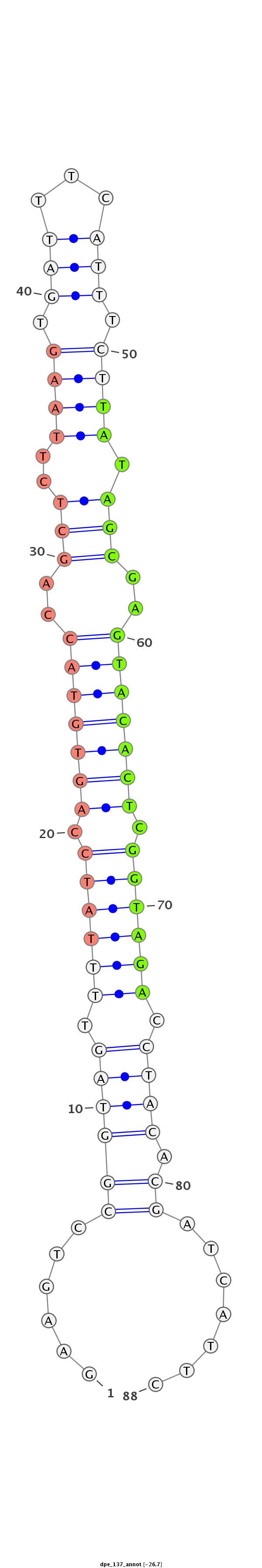
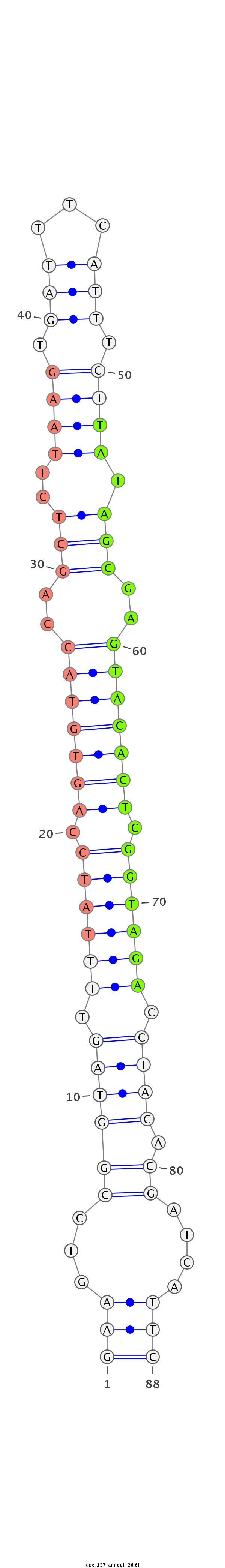
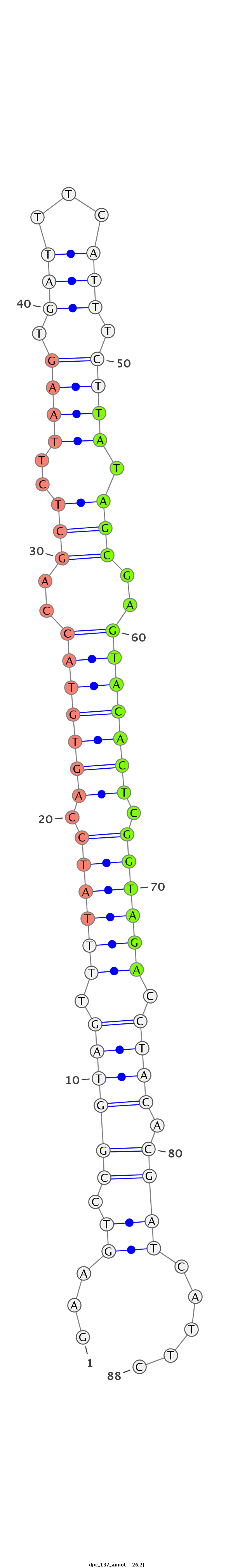
ID:dpe_137 |
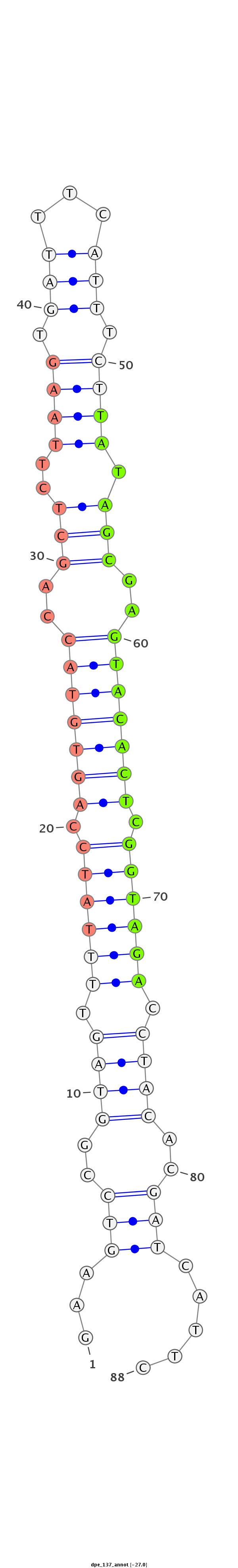
Coordinate:scaffold_13:191103-191176 + |
Confidence:candidate |
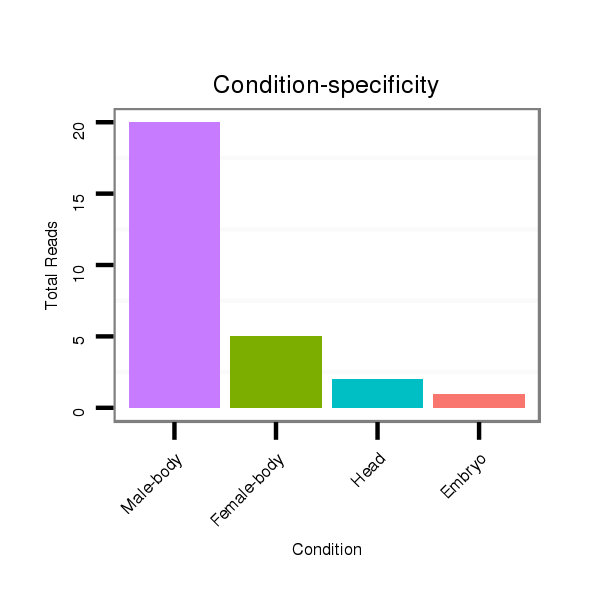
Class:Testes-restricted |
Genomic Locale:intergenic |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -26.7 | -26.6 | -26.2 |
 |
 |
 |
intergenic
No Repeatable elements found
|
CAAGACCAGCCGTGGGCGAAAAGACCCCGGAGGACAATCATTGGAAGTCCGGTAGTTTTATCCAGTGTACCAGCTCTTAAGTGATTTCATTTCTTATAGCGAGTACACTCGGTAGACCTACACGATCATTCTCCATCATCGGTCAAGGATCAATACTGTTCCATTTGGTGTGTG
*******************************************...(((..((((.((((((.(((((((..(((..((((.(((...))).)))).)))..))))))).)))))).))))..))).....******************************************* |
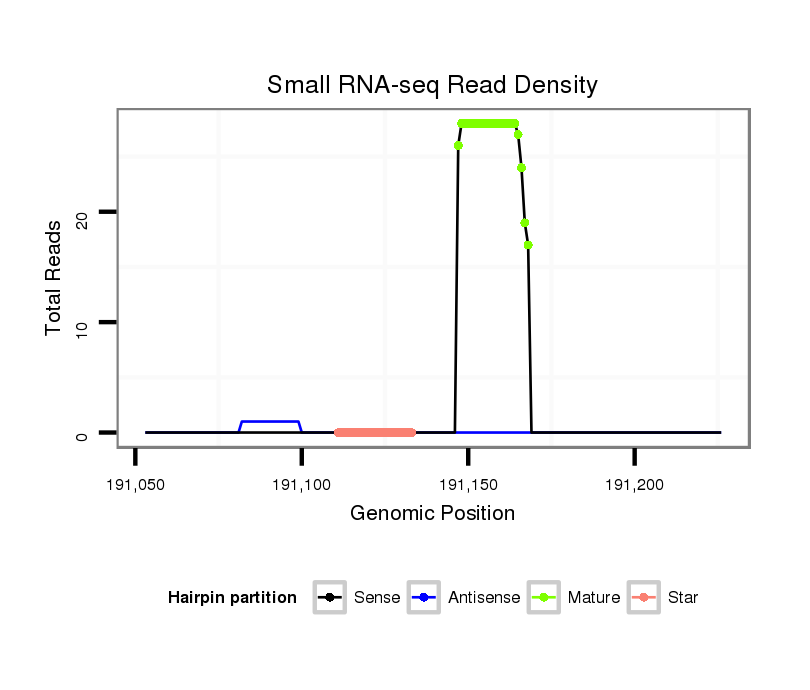
Read size | # Mismatch | Hit Count | Total Norm | Total | V111 male body |
M042 female body |
V050 head |
V057 head |
M021 embryo |
V042 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
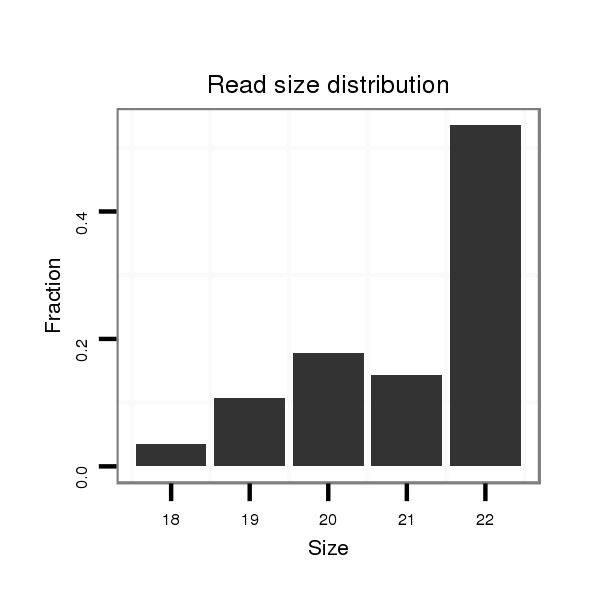
| ..............................................................................................TATAGCGAGTACACTCGGTAGA.......................................................... | 22 | 0 | 1 | 15.00 | 15 | 12 | 2 | 1 | 0 | 0 | 0 |
| ..............................................................................................TATAGCGAGTACACTCGGTA............................................................ | 20 | 0 | 1 | 5.00 | 5 | 4 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................TATAGCGAGTACACTCGGT............................................................. | 19 | 0 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................TATAGCGAGTACACTCGGTAG........................................................... | 21 | 0 | 1 | 2.00 | 2 | 0 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................ATAGCGAGTACACTCGGTAGA.......................................................... | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................TATAGCGAGTACAATCGGTAGA.......................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................ATAGCGAGTACACTCGGTAGC.......................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TATCCAGTGTACCAGCTCTTC............................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................TATAGCGAGTACACTCGG.............................................................. | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................TTTAGCGAGTACACTCGGTAG........................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................ATAGCGAGGACACTCGGTAG........................................................... | 20 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................TATAGCGAGTACACTCGGTAGC.......................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................TATCCAGTGTACCAGCTCTTAAGA............................................................................................ | 24 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................TATAGCGAGTACACTCGGTAGTCC........................................................ | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................TGCAAGTACGGTCGTTTTA.................................................................................................................. | 19 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........CGTGGGGGAAACGACCCAG................................................................................................................................................. | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................GATCAATACTGAGCCACTT........ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GTTCTGGTCGGCACCCGCTTTTCTGGGGCCTCCTGTTAGTAACCTTCAGGCCATCAAAATAGGTCACATGGTCGAGAATTCACTAAAGTAAAGAATATCGCTCATGTGAGCCATCTGGATGTGCTAGTAAGAGGTAGTAGCCAGTTCCTAGTTATGACAAGGTAAACCACACAC
*******************************************...(((..((((.((((((.(((((((..(((..((((.(((...))).)))).)))..))))))).)))))).))))..))).....******************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M021 embryo |
V057 head |
|---|---|---|---|---|---|---|---|
| ...................................................................................................................TGGATTTGCTAGTAAGAGGTA...................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 |
| .............................CTCCTGTTAGTAACCTTC............................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| .............................................................................................................................ATTAAGAGGTAGTGGGCAGT............................. | 20 | 3 | 12 | 0.08 | 1 | 0 | 1 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droPer2 | scaffold_13:191061-191219 + | dpe_137 | candidate | GCCGTGGGCGAAAAGACCCCGGAGGACAATCATTGGAAGTCCGGTAGTTTTATCCAGTGTACCAGCTCTTAAGTGATTTCATTTCTTATAGCGAGTACACTCGGTAGACCTACACGATCATTCTCCATCATCGGTCAAGGATCAATACTGTTCCATTTG |
| dp5 | XL_group1e:8373149-8373307 - | dps_3833 | candidate | GCCGTGTACGAAAAGACCCCGGACGACAATCATTGGAAGTCTGGTACTTTTATCCAGTGTACCAGCTCTTAAGTGATCTCATTTCTTATAGCGAGTACACTCGGTAGACCTACACGTTCATTCTCCATCATCGGTCAAGGGTCAATACTGTTCCCTTTG |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
Generated: 10/20/2015 at 06:48 PM