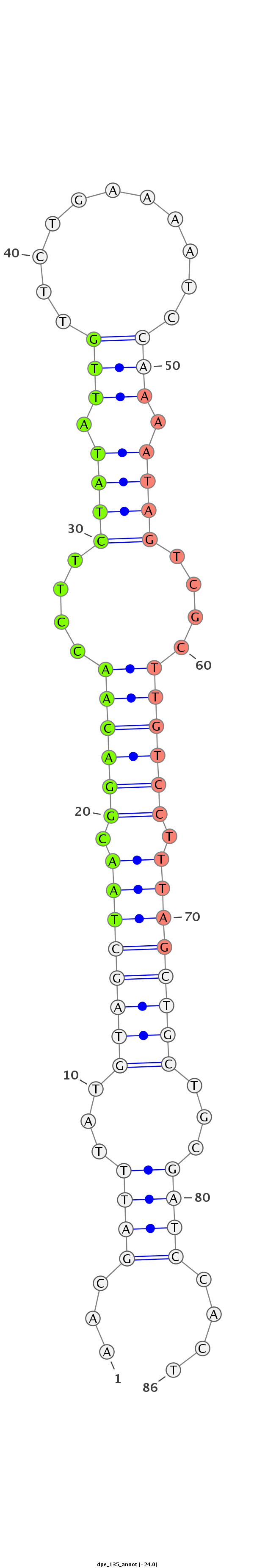
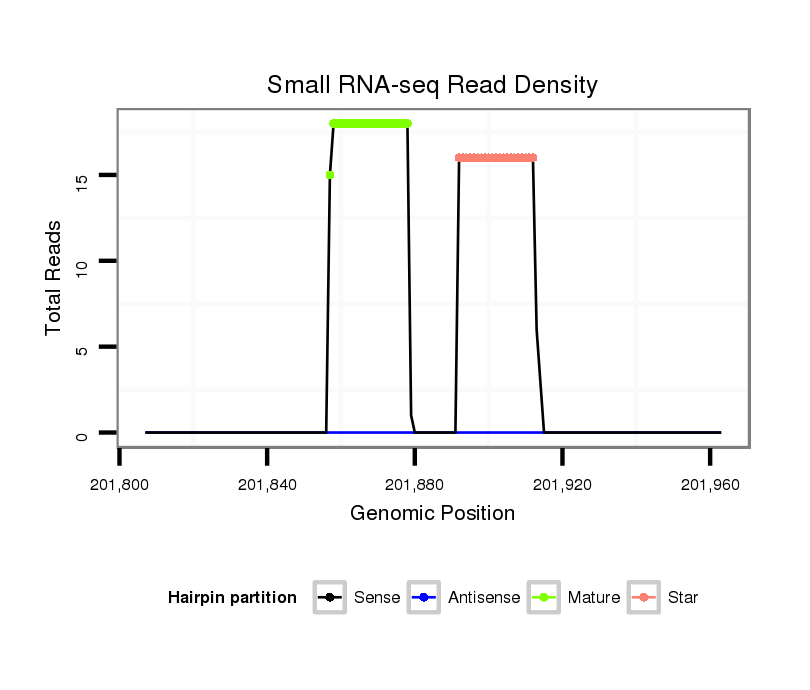
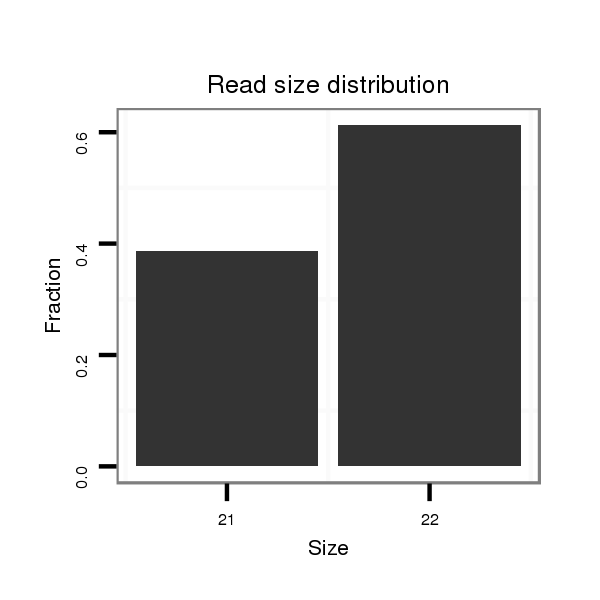
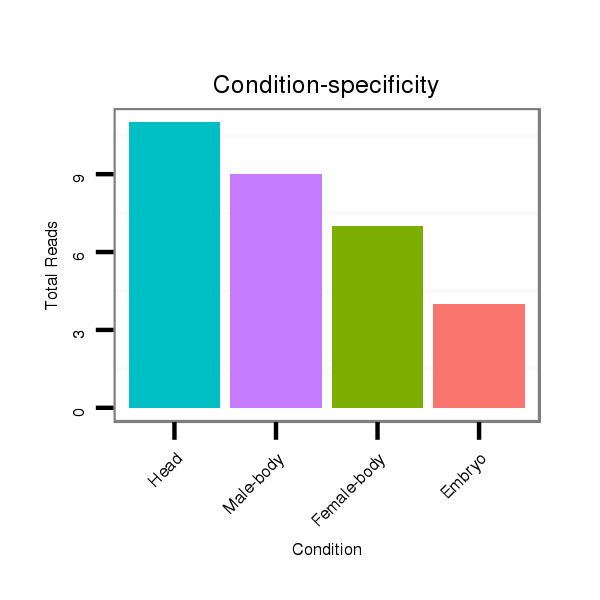
ID:dpe_135 |
Coordinate:scaffold_13:201857-201913 + |
Confidence:Known Ortholog |
Class:Testes-restricted |
Genomic Locale:intergenic |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
intergenic
No Repeatable elements found
|
TGCAATCAAAAAAAGCATCGCATTCCAGAATCATAAACGATTTATGTAGCTAACGGACAACCTTCTATATTGTTCTGAAAATCCAAAATAGTCGCTTGTCCTTTAGCTGCTGCGATCCACTTGGCAACGATCGAGCACGAACCGTCGACGATCGGTT
***********************************...((((...((((((((.((((((....((((.(((...........))).))))....)))))).))))))))...))))....************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | V111 male body |
V057 head |
M042 female body |
V050 head |
M021 embryo |
V042 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................TAACGGACAACCTTCTATATTG..................................................................................... | 22 | 0 | 1 | 15.00 | 15 | 3 | 4 | 4 | 2 | 2 | 0 |
| .....................................................................................AAATAGTCGCTTGTCCTTTAG................................................... | 21 | 0 | 1 | 10.00 | 10 | 6 | 1 | 0 | 3 | 0 | 0 |
| .....................................................................................AAATAGTCGCTTGTCCTTTAGC.................................................. | 22 | 0 | 1 | 3.00 | 3 | 0 | 1 | 1 | 0 | 0 | 1 |
| ..................................................TAACGGACAACCTTCTATATTGA.................................................................................... | 23 | 1 | 1 | 3.00 | 3 | 2 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................AAATAGTCGCTTGTCCTTTAGCT................................................. | 23 | 0 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 | 0 | 0 |
| ...................................................AACGGACAACCTTCTATATTG..................................................................................... | 21 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| ...................................................AACGGACAACCTTCTATATTGT.................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................AAATAGTCGCTTTTCCTTTAG................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................AAATAGTCGCTTGTCCTTTAGCC................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................AAATAGTCGCTTGTCCTTTCGC.................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................AAATAGTCGCTTGTCCATTAG................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................AAATAGCCGCTTGTCCTTTAGC.................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| TGCAATGTAAATAAGCATCGCA....................................................................................................................................... | 22 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................ACAACCTTCTTGATTGTGCTG................................................................................ | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| TGGCATCAAAAGAAGCATCGC........................................................................................................................................ | 21 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
|
ACGTTAGTTTTTTTCGTAGCGTAAGGTCTTAGTATTTGCTAAATACATCGATTGCCTGTTGGAAGATATAACAAGACTTTTAGGTTTTATCAGCGAACAGGAAATCGACGACGCTAGGTGAACCGTTGCTAGCTCGTGCTTGGCAGCTGCTAGCCAA
************************************...((((...((((((((.((((((....((((.(((...........))).))))....)))))).))))))))...))))....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M021 embryo |
V111 male body |
V057 head |
M042 female body |
V042 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| ....................GTAAGGTCTTATTATTTG....................................................................................................................... | 18 | 1 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................CGAGCTTGGCAGCTGTTGG.... | 19 | 3 | 15 | 1.00 | 15 | 5 | 6 | 2 | 1 | 1 |
| ....................................................................................................................................CCCGAGCTTGGCAGCTGTT...... | 19 | 3 | 10 | 0.50 | 5 | 3 | 0 | 1 | 1 | 0 |
| .............................................CATCGATTGCCAGTGGGC.............................................................................................. | 18 | 3 | 17 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droPer2 | scaffold_13:201807-201963 + | dpe_135 | confident | TGCAATCAAAAAAAGCATCGCATTCCAGAATCATAAACGATTTATGTAGCTAACGGACAACCTTCTATATTGTTCTGAAAATCCAAAATAGTCGCTTGTCCTTTAGCTGCTGCGATCCACTTGGCAACGATCG----------------AGCACGAACCGTCGACGATCGGTT |
| dp5 | XL_group1e:8362453-8362625 - | dps-mir-2537 | Known_mirbase | TGCAATCAAAAAAAGCATCGCATACCAGAATCATGAACGATTTATGTAGCTAACGGACAACCGTCTATATTGTTCTAAAAATCCAAAATAGTCGCTTGTCCTTTAGCCGCTGCGATCCACTTGCCAACGATCGACTAACAATCCCAGGGAGAACGATCCGTCGACGATCGGTT |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
Generated: 10/20/2015 at 06:49 PM