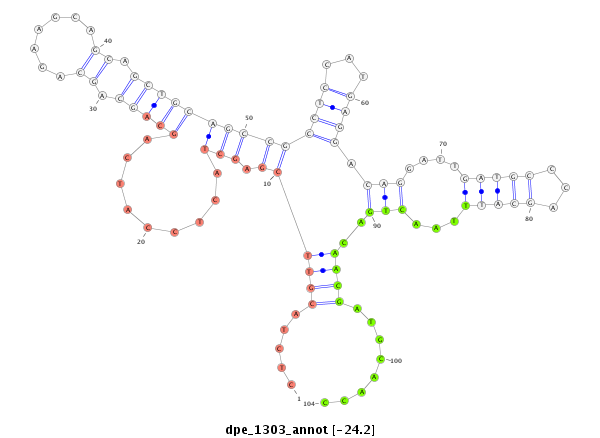
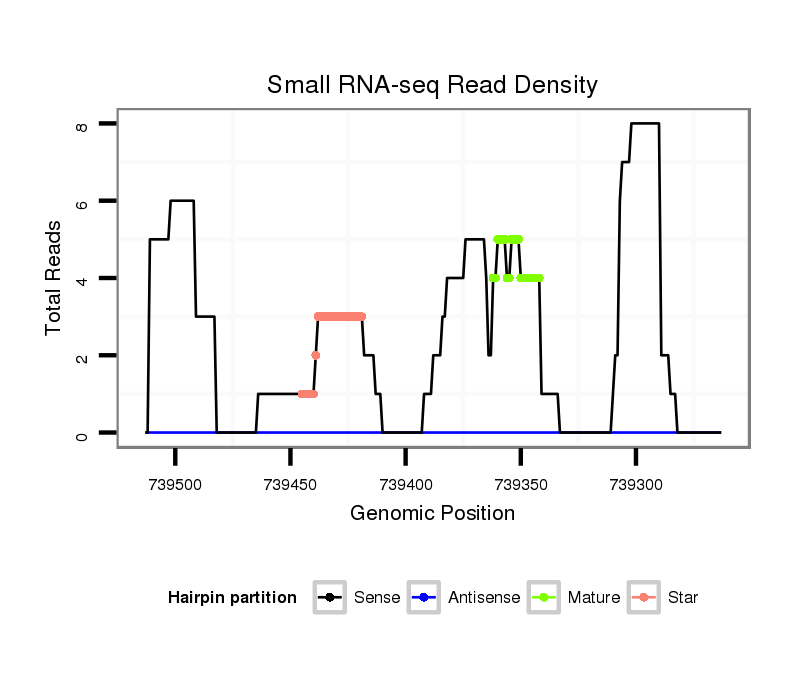
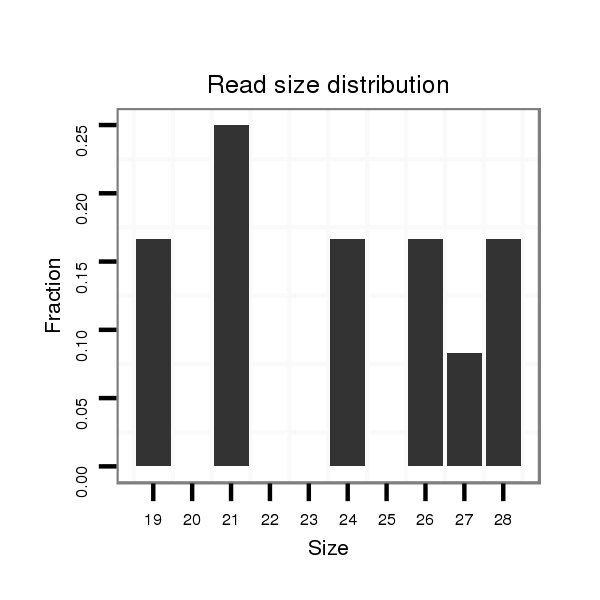
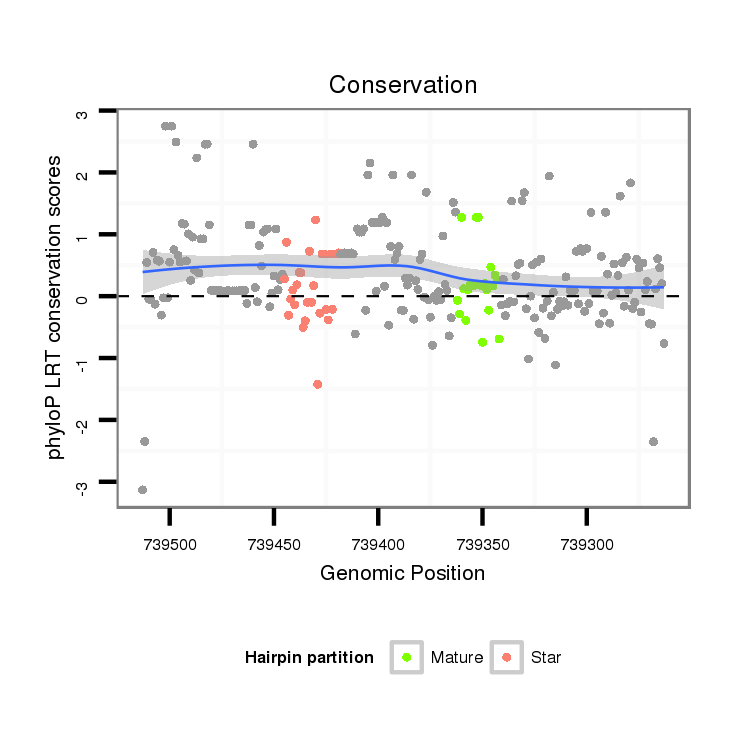
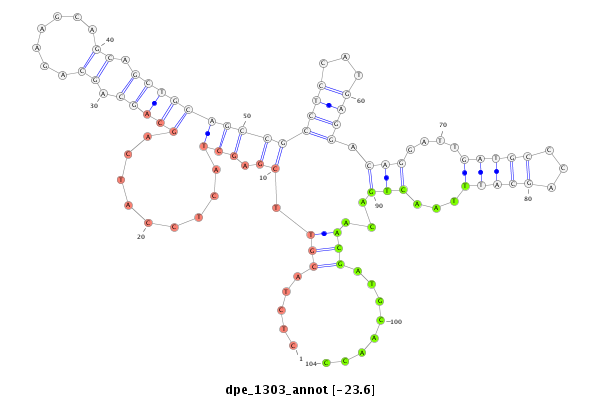
| TT----------------------------------------------------------------------------C----GCACTCTGATGAACAA--AA--TCACCCTGGAAATATATGCCACCGCCAAAGTAA-----TTG------TAATGCTTTCTCTCTACGTTCGAGCTAC---TCCATCAGCAGCAGCAGAAGCAGCAGCTGCAGC-----------------CGCCTCCATGAG------GAC--AGG-----------ATT-GATGCCCAGCATTTAACTGACAACGATGCAA----CCGCCACAGCCACATCCACAGCCTCAGCCA--CAGC--CACAGCCACACTTGAAT----------GCTGAAGCTGACTAGAAT-AA--TATCATTTC----------AGTTGTG | Size | Hit Count | Total Norm | Total | M022
Male-body | M040
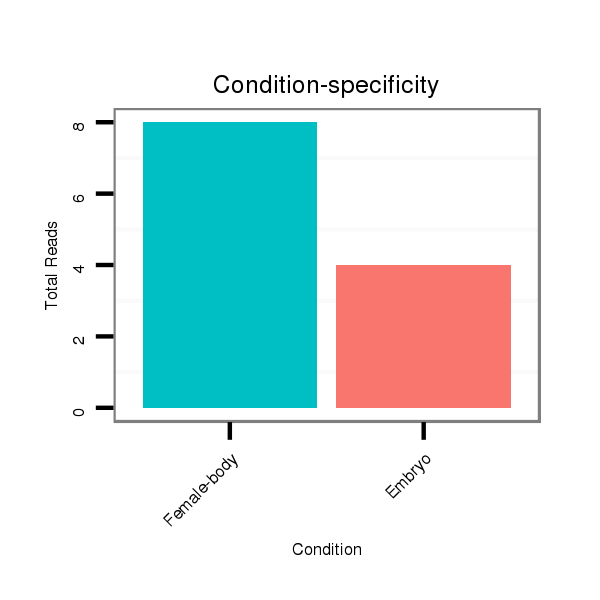
Female-body | M059
Embryo | M062
Head | V043
Embryo | V051
Head | V112
Male-body | SRR902010
Ovary | SRR902011
Testis | SRR902012
CNS imaginal disc |
| .............................................................................................................................................................................................................................................TCCATGAG------GAC--AGG-----------ATT-GATGCC............................................................................................................................................... | 23 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................AACTGACAACGATGCAA----CCGCC............................................................................................................. | 22 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................ACACTTGAAT----------GCTGAAGCT........................................ | 19 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................GCACTCTGATGAACAA--AA--TCACCCTGGA.................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................TAA-----TTG------TAATGCTTTCTCTCTAC.............................................................................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................CACTTGAAT----------GCTGAAGCTGACTAGA................................. | 25 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................C----GCACTCTGATGAACAA--AA--TCACCCT....................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................ACAGCCACATCCACAGCCTCAGCC..................................................................................... | 24 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................CTACGTTCGAGCTAC---TCCATCAGCA...................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................AAT----------GCTGAAGCTGACTAG.................................. | 18 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................AT----------GCTGAAGCTGACTAGA................................. | 18 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................GCAGC-----------------CGCCTCCATGAG------GAC--AGG-----------ATT-..................................................................................................................................................... | 26 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................GATGCCCAGCATTTAACTGACAACGATGC........................................................................................................................ | 29 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................TTCGAGCTAC---TCCATCAGCAGC.................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................TGCTTTCTCTCTACGTTCGAGCTAC---TCC............................................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................CTGATGAACAA--AA--TCACCCTGGA.................................................................................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................TGAAT----------GCTGAAGCTGACTAGAAT-AA--.......................... | 25 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................................................................CCATCAGCAGCAGCAGAAGC........................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................T----------GCTGAAGCTGACTAGAAT-A............................. | 20 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................ACTCTGATGAACAA--AA--TCACCCTG...................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................CTCCATGAG------GAC--AGG-----------ATT-GATGCCCAGC........................................................................................................................................... | 28 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................GCCTCCATGAG------GAC--AGG-----------ATT-G.................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................A-----TTG------TAATGCTTTCTCTCTACGTT........................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................................................AGCCACACTTGAAT----------GCTGAAGCT........................................ | 23 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................TCCATGAG------GAC--AGG-----------ATT-GATGCCCAGCA.......................................................................................................................................... | 28 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................CACATCCACAGCCTCAGCCA--CAGC--CAC......................................................................... | 27 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................ATGAACAA--AA--TCACCCTGGA.................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................CGAGCTAC---TCCATCAGCAGC.................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................GCCTCCATGAG------GAC--AGG-----------ATT-..................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................CACACTTGAAT----------GCTGAAGCT........................................ | 20 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................TCGAGCTAC---TCCATCAGCAGCAGC................................................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................CTCTACGTTCGAGCTAC---TCCATCAGCA...................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |