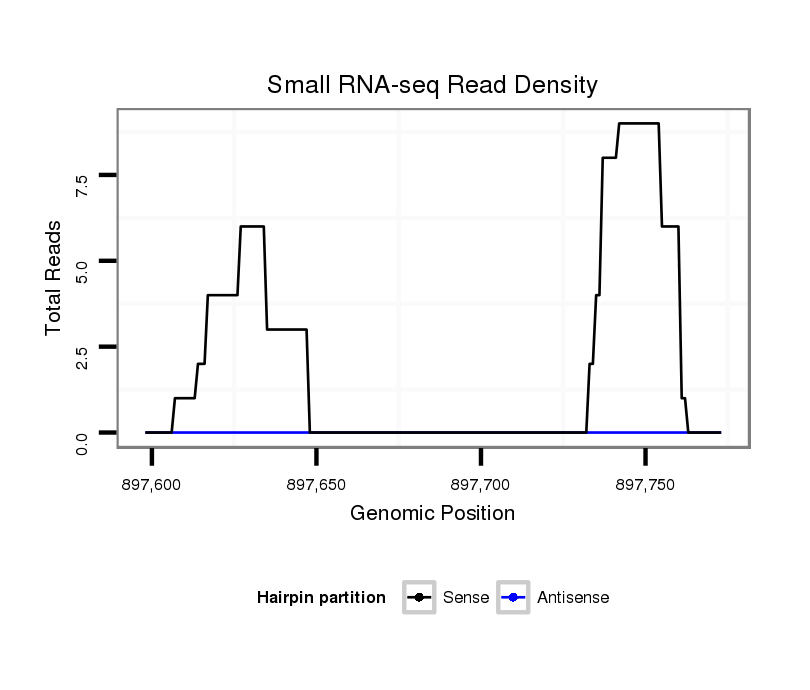
ID:dpe_1265 |
Coordinate:scaffold_25:897648-897723 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dper\GL26883-cds]; exon [dper_GLEANR_9417:1]; CDS [Dper\GL26883-cds]; exon [dper_GLEANR_9417:2]; intron [Dper\GL26883-in]
No Repeatable elements found
| --------------------------------------------######----------------------------------------------------------------------------################################################## TGTGTGTATGTTTAACTATTCTGGAAAAAATAAGAACGAACGCAATGAACGTAAGTATATCAACGCGAAACAGCACGTTTTTCCTGTTTGGTTTCACTAATTTGAAATACCTTCAATTGTTAAAAGGGAAAATTGGTGAAGCTCGATTCTGCCGACTGTGAAGGCTCATCTGATGA **************************************************....((((.((((.((.((((((..........)))))).)).........)))).))))................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M021 embryo |
V111 male body |
M042 female body |
|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................AGCTCGATTCTGCCGACT................... | 18 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 |
| .............................ATAAGAACGAACGCAATGAAC.............................................................................................................................. | 21 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 |
| .........................................................................................................................................GAAGCTCGATTCTGCCGACTGTGAAG............. | 26 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| ...................TCTGGAAAAAATAAGAAC........................................................................................................................................... | 18 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| .......................................................................................................................................GTGAAGCTCGATTCTGCCGACTGTGAAG............. | 28 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| ................TATTCTGGAAAAAATAAGAAC........................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .........................................................................................................................................GAAGCTCGATTCTGCCGACTGGGAAG............. | 26 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ................................................................................................................................................GATTCTGCCGACTGTGAAGGC........... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..........................................................................................................................AAAGGGCTAATTGGTGAGGCT................................. | 21 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| .........GTTTAACTATTCTGGAAAAA................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...........................................................................................................................................AGCTCGATTCTGCCGACTGTGAAG............. | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..............................................................................TTTTTCTGTTTGGATACACTA............................................................................. | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 |
| ..................................................................GAAACAGCACGTTTGTGA............................................................................................ | 18 | 3 | 20 | 0.10 | 2 | 0 | 2 | 0 |
| .............................................TGAACTTAAGCATATCAA................................................................................................................. | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 |
|
ACACACATACAAATTGATAAGACCTTTTTTATTCTTGCTTGCGTTACTTGCATTCATATAGTTGCGCTTTGTCGTGCAAAAAGGACAAACCAAAGTGATTAAACTTTATGGAAGTTAACAATTTTCCCTTTTAACCACTTCGAGCTAAGACGGCTGACACTTCCGAGTAGACTACT
**************************************************....((((.((((.((.((((((..........)))))).)).........)))).))))................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M042 female body |
V057 head |
V111 male body |
|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................GAGATAAGACGGCTGACAGT............... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..........................TTTGATAGTTGCTTGCGTTAC................................................................................................................................. | 21 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 |
| ..............................................................................AAAAAGACAAACCTATGTGAT............................................................................. | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droPer2 | scaffold_25:897598-897773 + | dpe_1265 | TGTGTGTATGTTTAACTATTCTGGAAAAAATAAGAACGAACGCAATGAACGTAAGTATATCAACGCGAAACAGCACGTTTTTCCTGTTTGGTTTCACTAATTTGAAATACCTTCAATTGTTAAAAGGGAAAATTGGTGAAGCTCGATTCTGCCGACTGTGAAGGCTCATCTGATGA |
| dp5 | XL_group1a:6423253-6423425 - | ---GTGTATGTTTAACTATTCTGGAAAAAATAAGAACGAACGCAATGAACGTAAGTATTTCAACGCGAGACAGCACGTTTTTCCTGTTTGGTTTCACTAATTTGAAATACCTTCAATTGTTAAAAGGGAAAATTGGTGAAGCTCGATTCTGCCGACTGTGAAGGCTCATCTGATGA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
Generated: 06/08/2015 at 03:56 PM