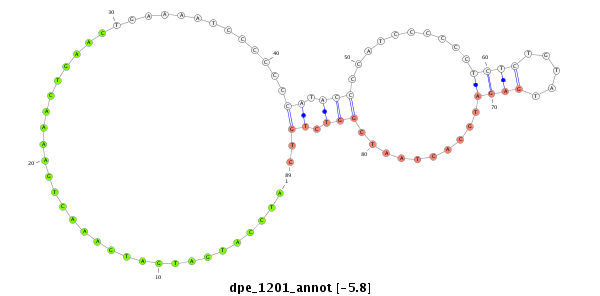
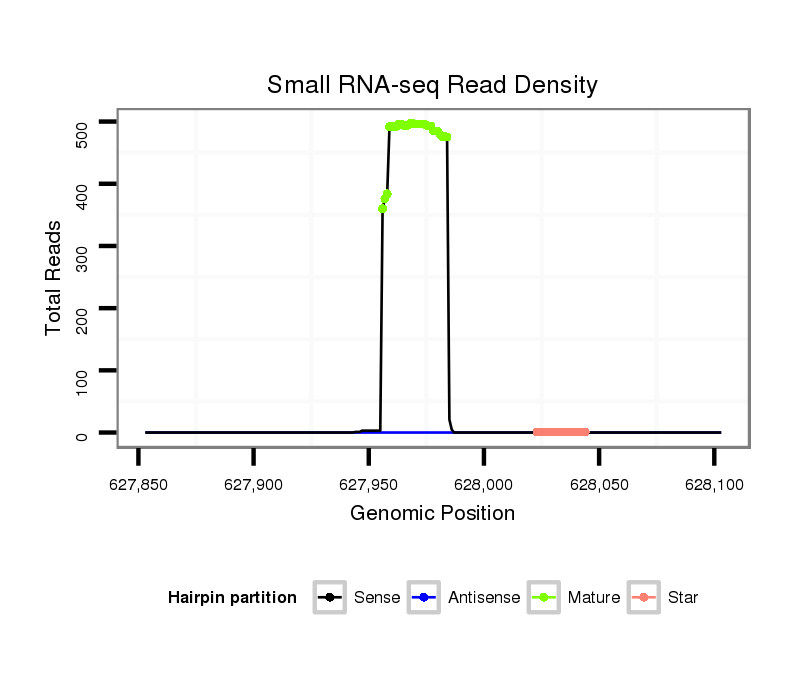
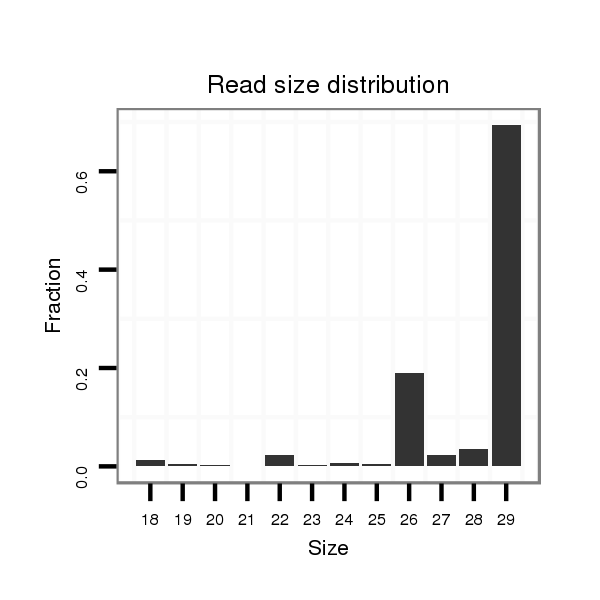
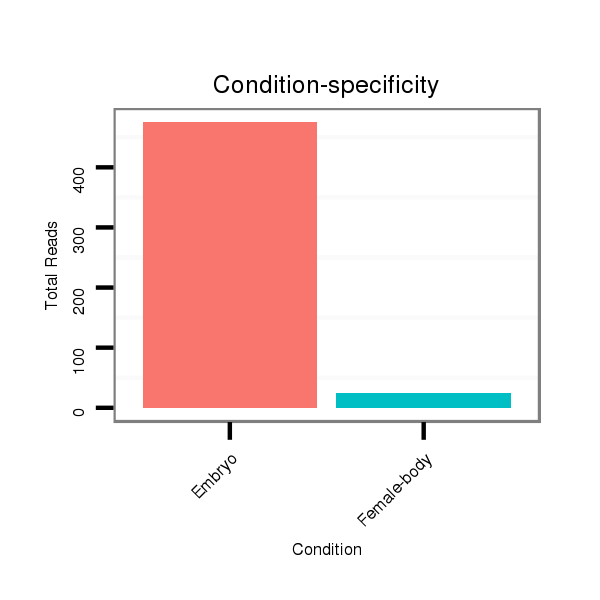
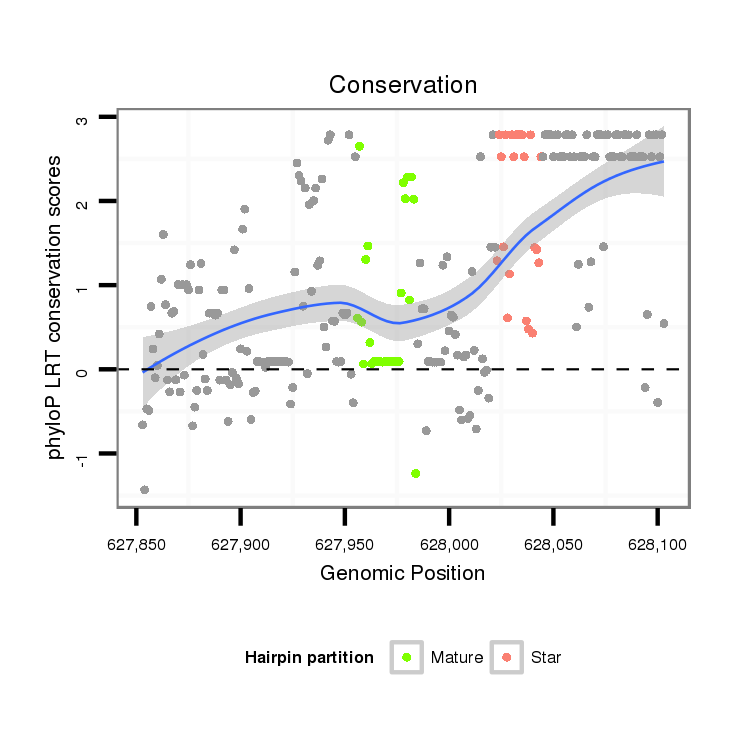
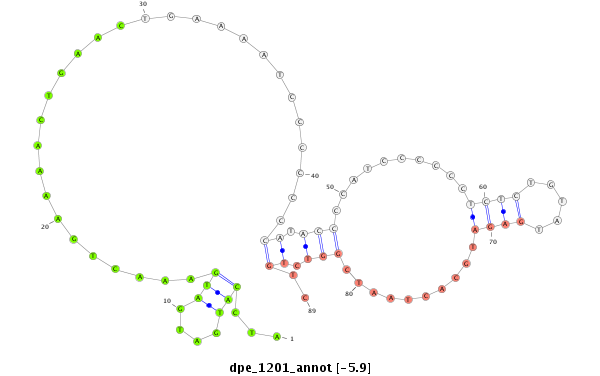
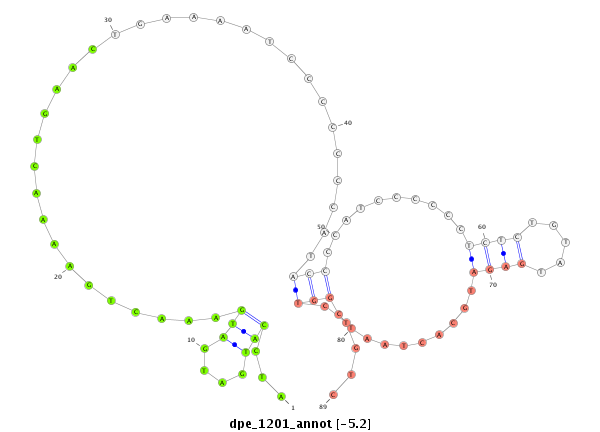
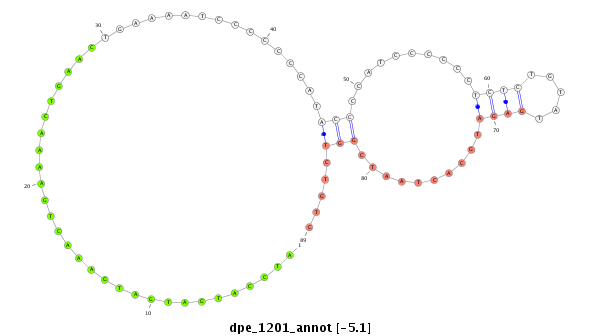
| .......................................................................................................ATCCATGATGATGAAACTGAAAACTGAAC....................................................................................................................... |
29 |
0 |
1 |
343.00 |
343 |
341 |
2 |
0 |
0 |
0 |
| ..........................................................................................................CATGATGATGAAACTGAAAACTGAAC....................................................................................................................... |
26 |
0 |
1 |
93.00 |
93 |
71 |
18 |
4 |
0 |
0 |
| ........................................................................................................TCCATGATGATGAAACTGAAAACTGAAC....................................................................................................................... |
28 |
0 |
1 |
12.00 |
12 |
12 |
0 |
0 |
0 |
0 |
| .......................................................................................................ATCCATGATGATGAAACTGAAACCTGAAC....................................................................................................................... |
29 |
1 |
1 |
10.00 |
10 |
10 |
0 |
0 |
0 |
0 |
| .......................................................................................................ATCCATGATGATGAAACTGAAA.............................................................................................................................. |
22 |
0 |
1 |
8.00 |
8 |
4 |
3 |
1 |
0 |
0 |
| ..........................................................................................................CATGATGATGAAACTGAAAACTGAACT...................................................................................................................... |
27 |
0 |
1 |
8.00 |
8 |
8 |
0 |
0 |
0 |
0 |
| ..........................................................................................................CATGATGATGAAACTGAAAACTGAACTG..................................................................................................................... |
28 |
0 |
1 |
5.00 |
5 |
5 |
0 |
0 |
0 |
0 |
| .......................................................................................................ATCCATGATGATTAAACTGAAAACTGAAC....................................................................................................................... |
29 |
1 |
1 |
5.00 |
5 |
5 |
0 |
0 |
0 |
0 |
| ........................................................................................................TCCATGATGATGAAACTGAAAACTGAACT...................................................................................................................... |
29 |
0 |
1 |
4.00 |
4 |
4 |
0 |
0 |
0 |
0 |
| .........................................................................................................CCATGATGATGAAACTGAAAACTGAAC....................................................................................................................... |
27 |
0 |
1 |
4.00 |
4 |
4 |
0 |
0 |
0 |
0 |
| ...................................................................................................................GAAACTGAAAACTGAACT...................................................................................................................... |
18 |
0 |
1 |
3.00 |
3 |
3 |
0 |
0 |
0 |
0 |
| ..............................................................................................................ATGATGAAACTGAAAACT........................................................................................................................... |
18 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
| .........................................................................................................CCATGATGATGAAACTGAAAACTG.......................................................................................................................... |
24 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
| ..............................................................................................................ATGATGAAACTGAAAACTGAAC....................................................................................................................... |
22 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
| .......................................................................................................ATCCATGATGATGAAACTG................................................................................................................................. |
19 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
| .......................................................................................................ATCCATGATGATGAAGCTGAAAACTGAAC....................................................................................................................... |
29 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
| ..............................................................................................GATGATGTGATCCATGAT........................................................................................................................................... |
18 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
| ..........................................................................................................CATGATGATGAAACTCAAAACTGAAC....................................................................................................................... |
26 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
| ..........................................................................................................CATGATGATAAAACTGAAAACTGAAC....................................................................................................................... |
26 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................................ATCCATGATGATGAAACTGA................................................................................................................................ |
20 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
| .........................................................................................................CCATGATGATGAAACTGAAAACT........................................................................................................................... |
23 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .........................................................................................................CCATGATGATGAAACTGAAAACTGAACT...................................................................................................................... |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................................ATCCATGATGATGAAACTGAAAACT........................................................................................................................... |
25 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................................ATCCATGATGATGAAACTGAAAAC............................................................................................................................ |
24 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................................TCCATGATGATGAAACTAAAAAC............................................................................................................................ |
23 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................................ATCCATGATGATGAAACTGAAAGCTGAAC....................................................................................................................... |
29 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................................ATCCATGATGATCAAACTGAAAACTGAAC....................................................................................................................... |
29 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..........................................................................................................CATGATGAGGAAACTGAAAACTGAAC....................................................................................................................... |
26 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ..........................................................................................................CATGATGATGAAACTGAGAACTGAAC....................................................................................................................... |
26 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..........................................................................................................................................................................GAGATGCACTAATCGGTCTGTC........................................................... |
22 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................................ATCCATGATGATGAACCTGAAAACTGAAC....................................................................................................................... |
29 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................................ATCCATGATGATGAAACTGAAAACTG.......................................................................................................................... |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................................ATCCATGAGGATGAAACTGAAAACTGAAC....................................................................................................................... |
29 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..........................................................................................................CATGATGATGAAACTGAAAACTGAA........................................................................................................................ |
25 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ..........................................................................................................CATGATGATGAAACTGAAAACTGCAC....................................................................................................................... |
26 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..........................................................................................................CATGATGATGAAACTGAAAACT........................................................................................................................... |
22 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..........................................................................................................CATGATGATGAACCTGAAAACTGAAC....................................................................................................................... |
26 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................................ATCCATGATTATGAAACTGAAAACTGAAC....................................................................................................................... |
29 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...........................................................................................GCAGATGATGTGATCCATGATGATGA...................................................................................................................................... |
26 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ..........................................................................................................CATGATGATGAAACTGAAAACTGGAC....................................................................................................................... |
26 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................................ATCCATGATGACGAAACTGAAAACTGAAC....................................................................................................................... |
29 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..........................................................................................................CATGATGATGAAACCGAAAACTGCAC....................................................................................................................... |
26 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................................................TGAACACAGAACTGAGAATCCC............................................................................................................. |
22 |
3 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
1 |
| .......................................................................................................ATCCATGATGATGAAACTGAAAATTGAAC....................................................................................................................... |
29 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..........................................................................................................CATGATTATGAAACTGAAAACTGAAC....................................................................................................................... |
26 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................................................TGAACACAGAACTGATAATCCC............................................................................................................. |
22 |
3 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
| .......................................................................................................ATCCATGATGATAAAACTGAAAACTGAAC....................................................................................................................... |
29 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..............GACAAGAACCCGGAACAG........................................................................................................................................................................................................................... |
18 |
2 |
3 |
0.67 |
2 |
0 |
2 |
0 |
0 |
0 |
| .............TGACAAGAACCCGGAACAG........................................................................................................................................................................................................................... |
19 |
3 |
20 |
0.15 |
3 |
0 |
3 |
0 |
0 |
0 |
| ..........................................................CTATAACTAAAAGTAAAATGAA........................................................................................................................................................................... |
22 |
3 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
0 |