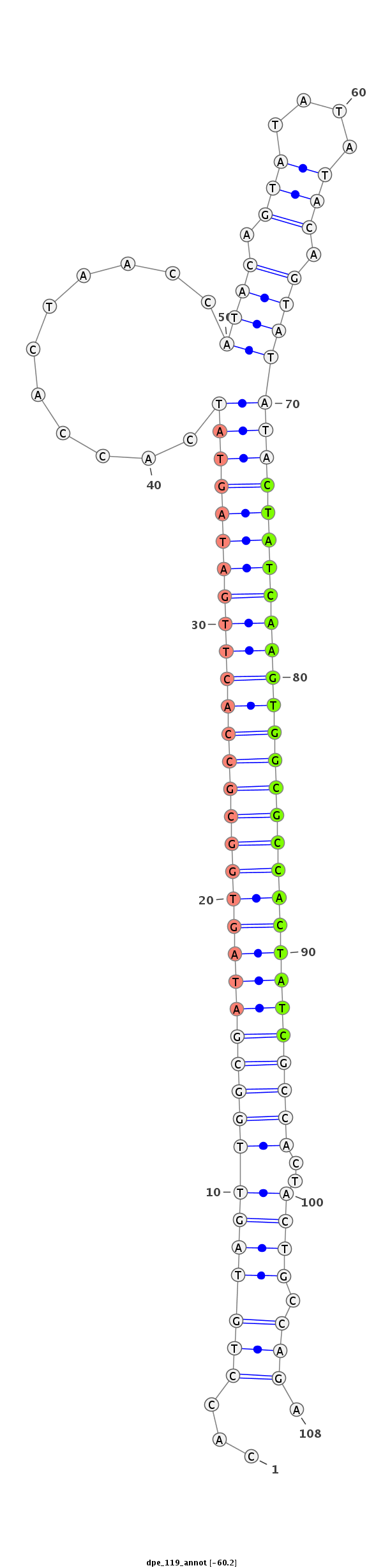
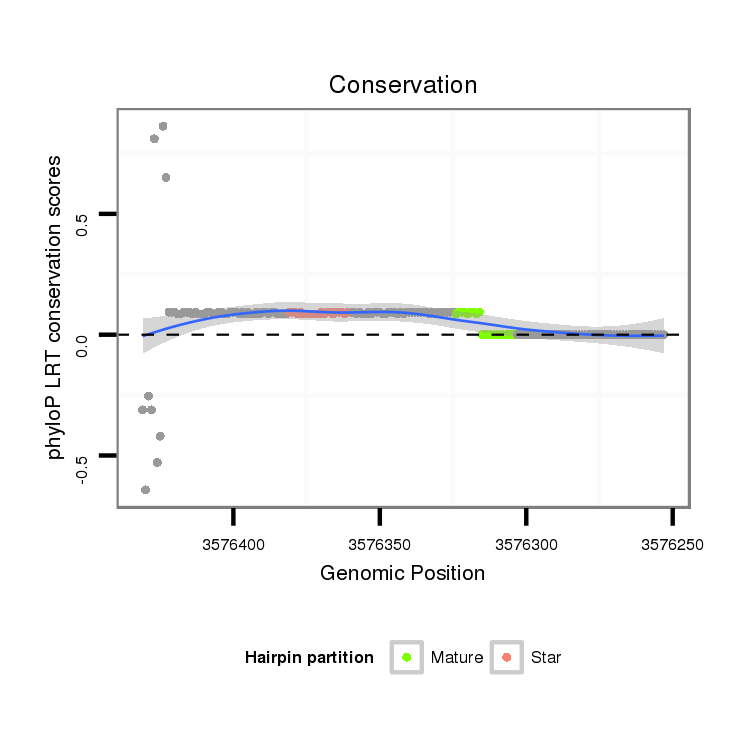
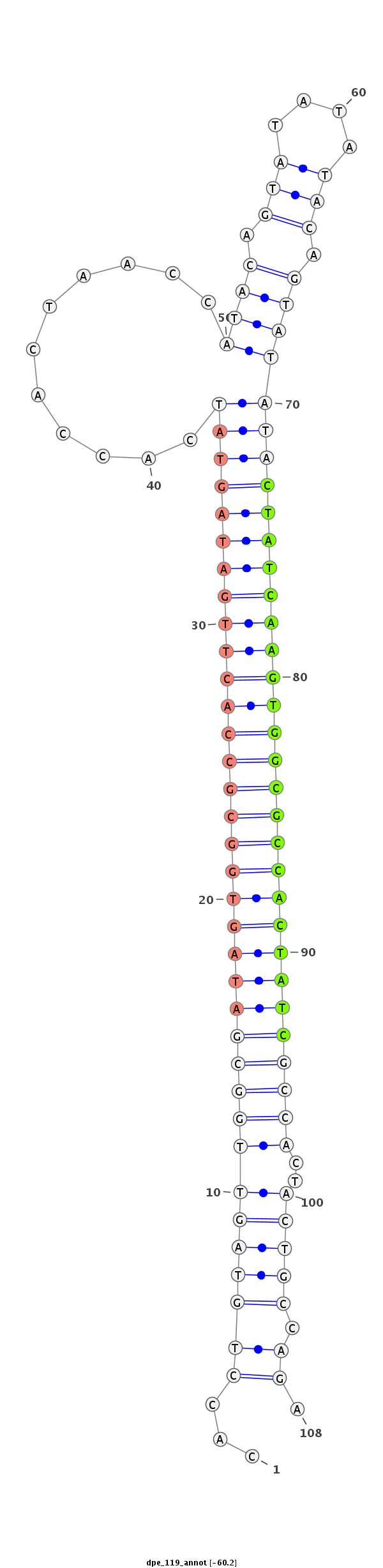
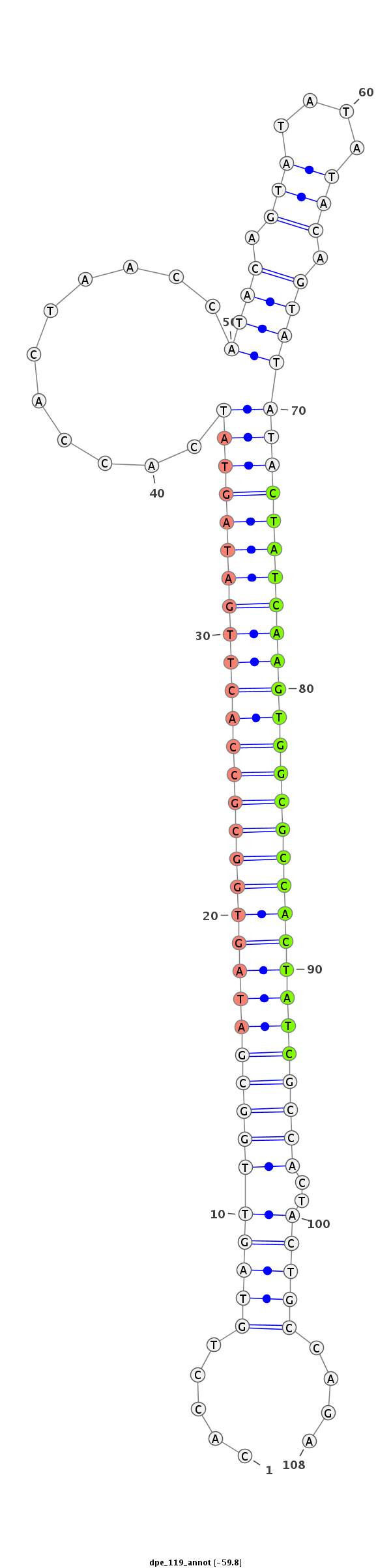
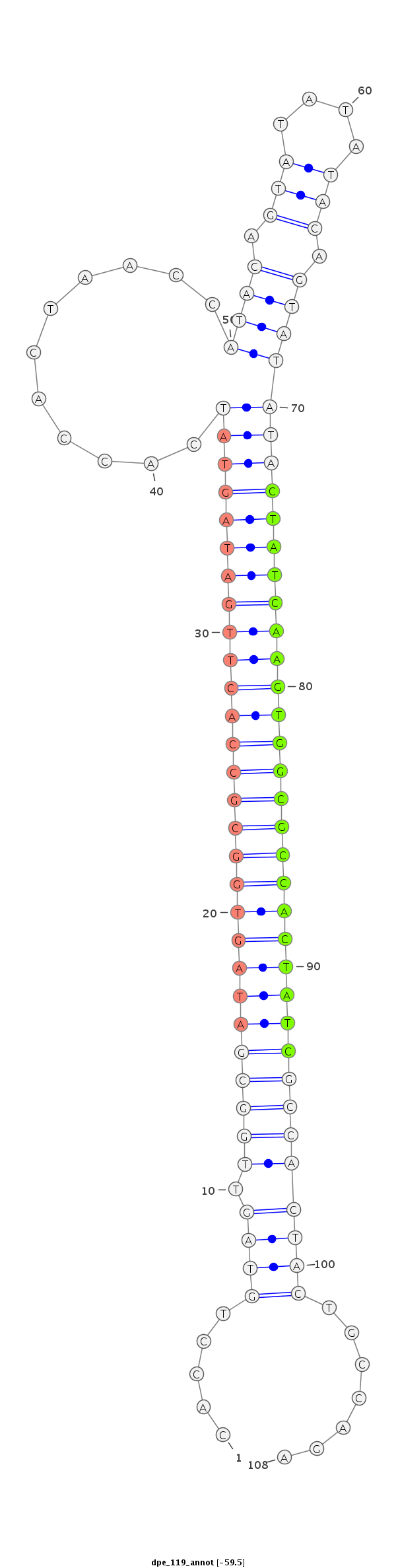
ID:dpe_119 |
Coordinate:scaffold_5:3576303-3576381 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
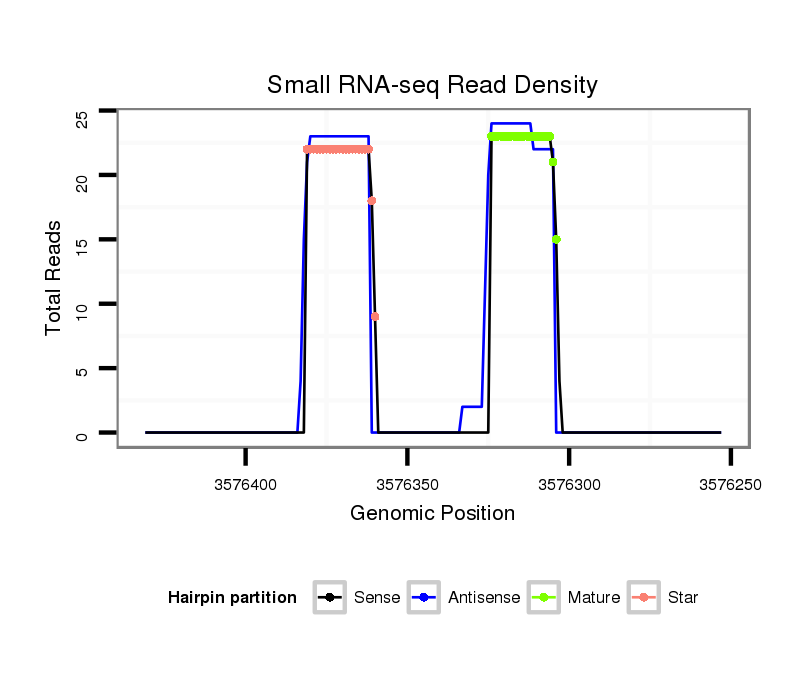
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -60.2 | -59.8 | -59.5 |
 |
 |
 |
intergenic
No Repeatable elements found
| mature | star |
|
TACAGTTACTATCGATAGTGCCATCGATGGTTTGACACCTGTAGTTGGCGATAGTGGCGCCACTTGATAGTATCACCACTAACCATACAGTATATATACAGTATATACTATCAAGTGGCGCCACTATCGCCACTACTGCCAGAGTGGCGCTGGGCTTGGGAAAATATCGAAATGATTTT
***********************************...(((((((((((((((((((((((((((((((((((...........((((.(((....))).))))))))))))))))))))))))))))))))..)))).))).************************************ |
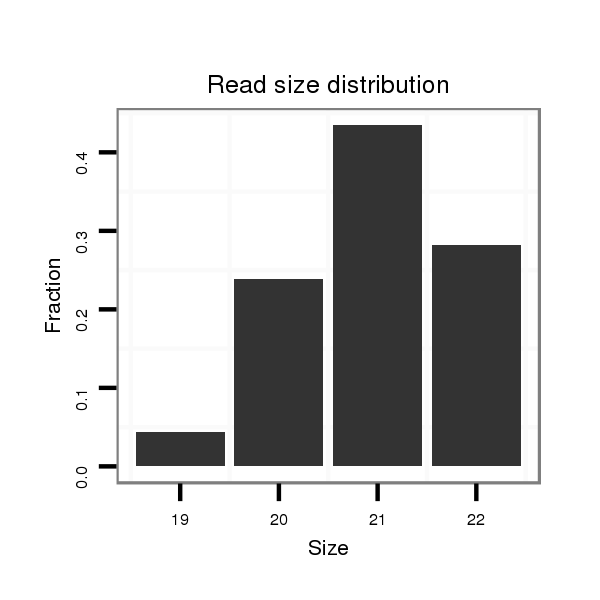
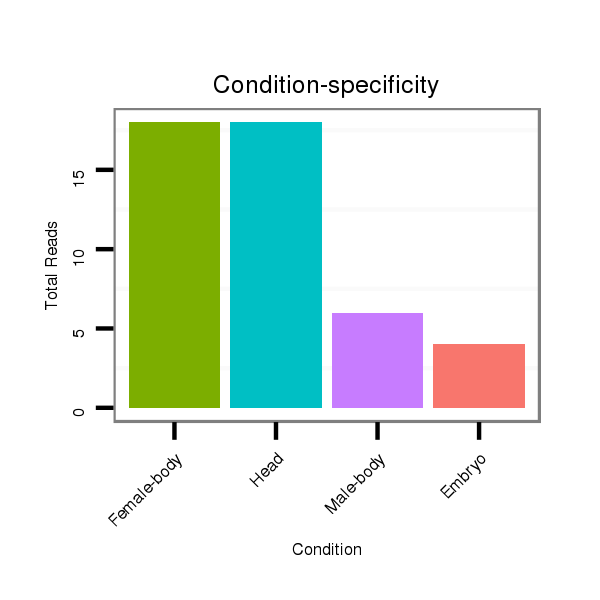
Read size | # Mismatch | Hit Count | Total Norm | Total | M042 female body |
V057 head |
V050 head |
V111 male body |
V042 embryo |
M021 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................CTATCAAGTGGCGCCACTATC................................................... | 21 | 0 | 2 | 11.00 | 22 | 6 | 5 | 3 | 2 | 6 | 0 |
| ..................................................ATAGTGGCGCCACTTGATAGTA........................................................................................................... | 22 | 0 | 2 | 9.00 | 18 | 7 | 3 | 5 | 1 | 2 | 0 |
| ..................................................ATAGTGGCGCCACTTGATAGT............................................................................................................ | 21 | 0 | 2 | 9.00 | 18 | 8 | 5 | 1 | 4 | 0 | 0 |
| ...........................................................................................................CTATCAAGTGGCGCCACTAT.................................................... | 20 | 0 | 2 | 6.00 | 12 | 5 | 4 | 2 | 1 | 0 | 0 |
| ..................................................ATAGTGGCGCCACTTGATAG............................................................................................................. | 20 | 0 | 2 | 4.50 | 9 | 0 | 3 | 2 | 4 | 0 | 0 |
| ...........................................................................................................CTATCAAGTGGCGCCACTATCG.................................................. | 22 | 0 | 2 | 4.00 | 8 | 6 | 2 | 0 | 0 | 0 | 0 |
| ...........................................................................................................CTATCAAGTGGCGCCACTA..................................................... | 19 | 0 | 2 | 2.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................CTGTCAAGTGGCGCCACTATC................................................... | 21 | 1 | 2 | 1.50 | 3 | 1 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................................CTATCAAGTGGCGCCACTATCGT................................................. | 23 | 1 | 2 | 1.50 | 3 | 2 | 1 | 0 | 0 | 0 | 0 |
| ..................................................ATAGTGGCGCCACTTGATAGTAA.......................................................................................................... | 23 | 1 | 2 | 1.00 | 2 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................ATAGTGGCGCCGCTTGATAGTA........................................................................................................... | 22 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ATAGTGGCCCCACTTGATAGT............................................................................................................ | 21 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TAGTGGCGCCACTTGATAGT............................................................................................................ | 20 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................TGTCAAGTGGCGCCACTATCG.................................................. | 21 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................CGATAGTGGCGCCACTTGA................................................................................................................ | 19 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................CTATCAAGTGGCGCCACTATCT.................................................. | 22 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ATAGTGGCGCCACTTGTTAGT............................................................................................................ | 21 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................ATAGTGGCGCCGCTTGATA.............................................................................................................. | 19 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ATAGTGGCGCCCCTTGATAGTA........................................................................................................... | 22 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................ATAGCGGCGCCACTTGATAGTA........................................................................................................... | 22 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................CTAGCAAGAGGCGCCACTATCC.................................................. | 22 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................CTGTCAGGTGGCGCCACTATCG.................................................. | 22 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................CTATCAAGTGGCGCCACTATA................................................... | 21 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................CTATCAGGTGGCGCCACTATC................................................... | 21 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ATAGTCGCGCCACTTGATAGT............................................................................................................ | 21 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................ATGGTGGCGCCACTTGATAGTA........................................................................................................... | 22 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ATAGTGGCGCCACTTGCTGGT............................................................................................................ | 21 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................CTATCAAGTGGCGCCACTTTC................................................... | 21 | 1 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................CTATCAAGGGGCGCCACTAT.................................................... | 20 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................TTGGCGATAGGGGCGACA..................................................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................ATAGTGGCGCCCCTTGCTAC............................................................................................................. | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................TTGGGGATAGTGGCGCAA..................................................................................................................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................ATTTGGCGATAGTGGCGCTT..................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............GATAGTGCCATCGATGGTTTGA................................................................................................................................................ | 22 | 0 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................ATTGGGGATAGTGGCGCCAG.................................................................................................................... | 20 | 3 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................TTGGCGATAGGGGCGTAA..................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................GGAAAACATCGACATGATTG. | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
|
ATGTCAATGATAGCTATCACGGTAGCTACCAAACTGTGGACATCAACCGCTATCACCGCGGTGAACTATCATAGTGGTGATTGGTATGTCATATATATGTCATATATGATAGTTCACCGCGGTGATAGCGGTGATGACGGTCTCACCGCGACCCGAACCCTTTTATAGCTTTACTAAAA
************************************...(((((((((((((((((((((((((((((((((((...........((((.(((....))).))))))))))))))))))))))))))))))))..)))).))).*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M042 female body |
V057 head |
V050 head |
V111 male body |
V042 embryo |
M021 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................CTATCACCGCGGTGAACTATC............................................................................................................. | 21 | 0 | 2 | 11.00 | 22 | 6 | 5 | 3 | 2 | 6 | 0 |
| ..............................................................................................................................AGCGGTGATGATCGTCTTA.................................. | 19 | 3 | 9 | 11.00 | 99 | 86 | 11 | 0 | 2 | 0 | 0 |
| ..........................................................................................................TGATAGTTCACCGCGGTGATA.................................................... | 21 | 0 | 2 | 9.00 | 18 | 8 | 5 | 1 | 4 | 0 | 0 |
| .........................................................................................................ATGATAGTTCACCGCGGTGATA.................................................... | 22 | 0 | 2 | 9.00 | 18 | 7 | 3 | 5 | 1 | 2 | 0 |
| ..................................................TATCACCGCGGTGAACTATC............................................................................................................. | 20 | 0 | 2 | 6.00 | 12 | 5 | 4 | 2 | 1 | 0 | 0 |
| ...........................................................................................................GATAGTTCACCGCGGTGATA.................................................... | 20 | 0 | 2 | 4.50 | 9 | 0 | 3 | 2 | 4 | 0 | 0 |
| ................................................GCTATCACCGCGGTGAACTATC............................................................................................................. | 22 | 0 | 2 | 4.00 | 8 | 6 | 2 | 0 | 0 | 0 | 0 |
| ...................................................ATCACCGCGGTGAACTATC............................................................................................................. | 19 | 0 | 2 | 2.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................GTCATATATGATAGTTCACCGC........................................................... | 22 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
| ...............................................................................................................................GCGGTGATGATCGTCTTA.................................. | 18 | 3 | 20 | 1.75 | 35 | 25 | 10 | 0 | 0 | 0 | 0 |
| .................................................CTATCACCGCGGTGAACTGTC............................................................................................................. | 21 | 1 | 2 | 1.50 | 3 | 1 | 1 | 1 | 0 | 0 | 0 |
| ...............................................TGCTATCACCGCGGTGAACTATC............................................................................................................. | 23 | 1 | 2 | 1.50 | 3 | 2 | 1 | 0 | 0 | 0 | 0 |
| ........................................CATCAACCACTATCACCTCGAT..................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................AATGATAGTTCACCGCGGTGATA.................................................... | 23 | 1 | 2 | 1.00 | 2 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................AGCGGTGATGATCGTCTT................................... | 18 | 3 | 20 | 0.90 | 18 | 15 | 3 | 0 | 0 | 0 | 0 |
| ..........................................................................TGGTGATTGGTATGTCATAT..................................................................................... | 20 | 0 | 20 | 0.55 | 11 | 0 | 11 | 0 | 0 | 0 | 0 |
| ..............................................................................................................AGTTCACCGCGGTGATAGC.................................................. | 19 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................ATATCACCGCGGTGAACTATC............................................................................................................. | 21 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................TGATAGTTCACCGCGGTGAT..................................................... | 20 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................TCTATCACCGCGGTGAACTATC............................................................................................................. | 22 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCTATCACCGCGGTGGACTGTC............................................................................................................. | 22 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................CTATCACCGCGGTGGACTATC............................................................................................................. | 21 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................ATGATAGTTCCCCGCGGTGATA.................................................... | 22 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................GCTATCACCGCGGTGAACTGT.............................................................................................................. | 21 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................TGATAGTTCACCGCGCTGATA.................................................... | 21 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................TGATTGTTCACCGCGGTGATA.................................................... | 21 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................CCTATCACCGCGGAGAACGATC............................................................................................................. | 22 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................ATGATAGTTCACCGCGGTGGTA.................................................... | 22 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................ATGATAGTTCGCCGCGGTGATA.................................................... | 22 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................ATGATAGTTCACCGCGGCGATA.................................................... | 22 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................ATAGTTCGCCGCGGTGATA.................................................... | 19 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................TGATAGTTCACCCCGGTGATA.................................................... | 21 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TATCACCGCGGGGAACTATC............................................................................................................. | 20 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................TGGTCGTTCACCGCGGTGATA.................................................... | 21 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................CTTTCACCGCGGTGAACTATC............................................................................................................. | 21 | 1 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................AAAGCGGTGATGAGGGTC..................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................CATCGTTCCCCGCGGTGATA.................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................AGCGGTGATGACCGTCTTAG................................. | 20 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................CCGCGGCGATAGCGGTAA............................................. | 18 | 2 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................TGGTGATTGGTATGTCATATA.................................................................................... | 21 | 0 | 20 | 0.10 | 2 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................GACCGCGGTGATAGGGGTTA............................................. | 20 | 3 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................ACCGCGACCCGCACCCAT................. | 18 | 2 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................TGATTGGTATGTCATATAT................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................TTGGTATGTTATATAAATGG............................................................................... | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droPer2 | scaffold_5:3576253-3576431 - | dpe_119 | TACAGTTACTATCGATAGTGCCATCGATGGTTTGACACCTGTAGTTGGCGATAGTGGCGCCACTTGATAGTATCACCACTAACCATACAGTATATATACAGTATATACTATCAAGTGGCGCCACTATCGCCACTACTGCCAGAGTGGCGCTGGGCTTGGGAAAATATCGAAATGATTTT |
| dp5 | 4_group1:1629860-1629975 + | TACAGTTACTATCGATAGTGCCATCGATGGTTTGACACCTGTAGTTGGCGATAGTGGCGCCACTTGATAGTATCACCACTAACCATACAGTATATATACAGTATATACTATCAAGT--------------------------------------------------------------- | |
| droTak1 | scf7180000415858:122780-122788 - | TCTAGTAAC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEre2 | scaffold_4929:15469620-15469627 + | AACTGGTA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 06/28/2015 at 04:05 PM