
ID:dpe_1168 |
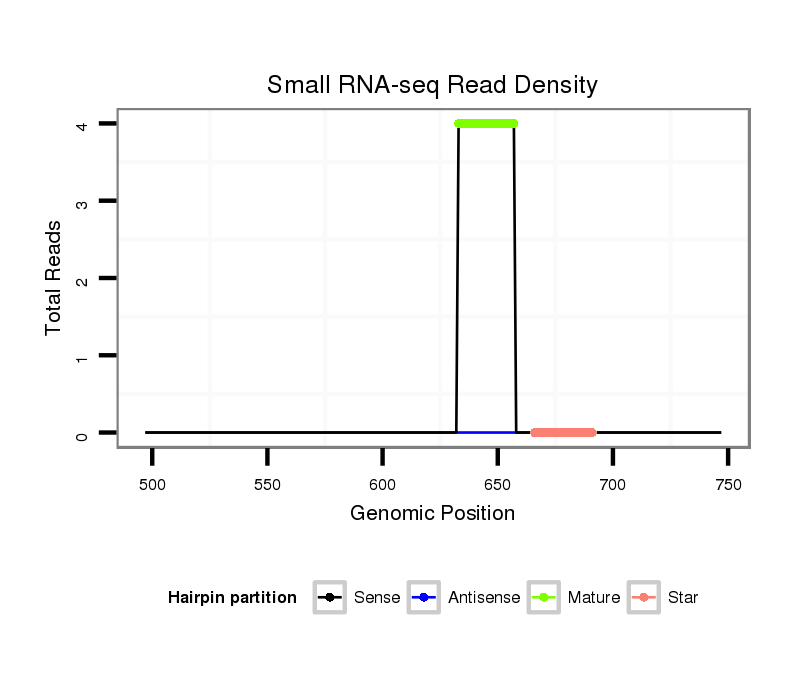
Coordinate:scaffold_2131:547-697 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
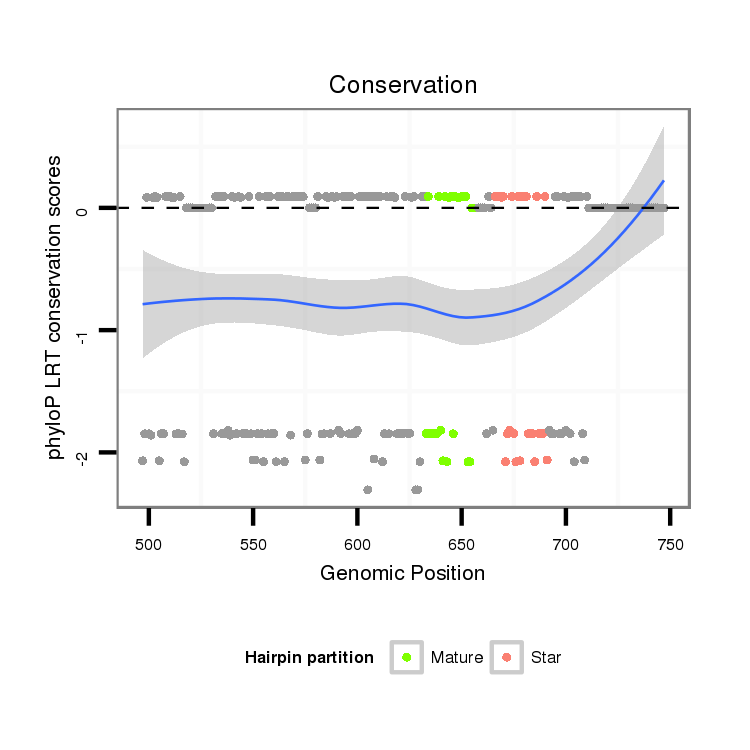
| -7.2 | -7.1 | -6.9 |
 |
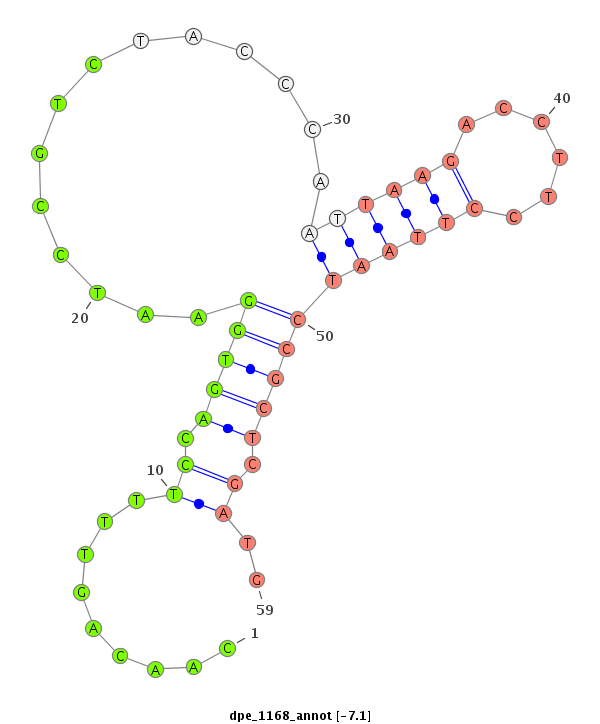 |
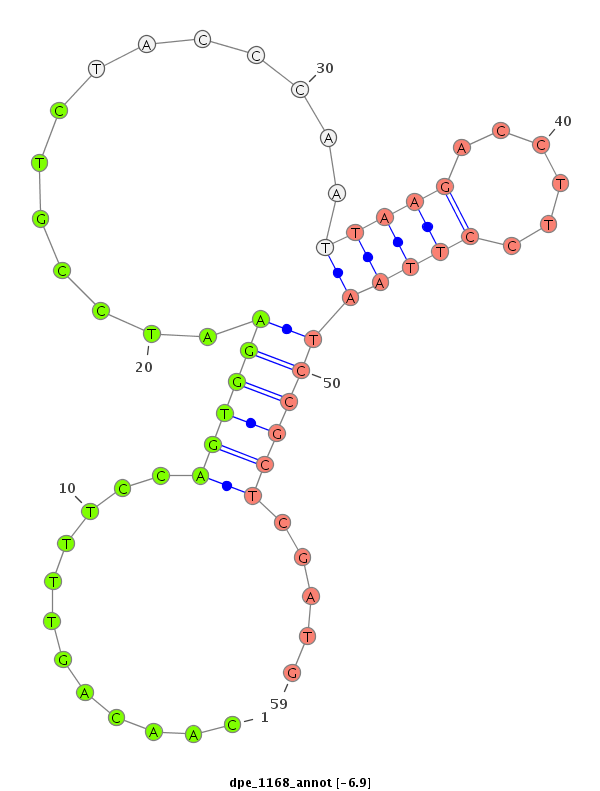 |
exon [dper_GLEANR_16538:1]; CDS [Dper\GL15395-cds]; intron [Dper\GL15395-in]
| Name | Class | Family | Strand |
| BEL3-LTR_Dpse | LTR | Pao | - |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TGCGTCTGTGCATTCCACAACTCGCCTTCTTGGGCTTACTCTTAGCACAGGTGGGCTCCTGAAACATTCTTTAAATATGCGGTAAGAGAGCTCATCTTAGACATTTTTCATAAAACCTGTACGAGCTAAGTCCCTTCAACAGTTTTCCAGTGGAATCCGTCTACCCAATTAAGACCTTCCTTAATCCGCTCGATGTAGTTCCTTGCTCATTCGTTTTATTTTGTATATCTCTTAAATCTAGTGATCATAAT ****************************************************************************************************************************************.........((.((((((..............(((((......))))))))))).))..******************************************************** |
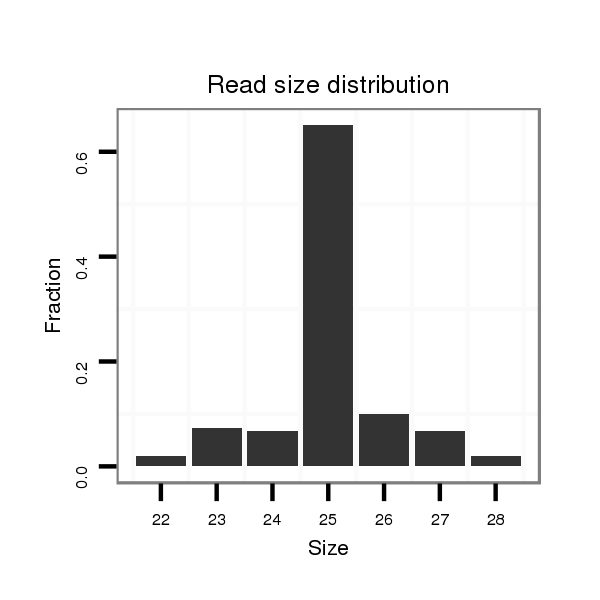
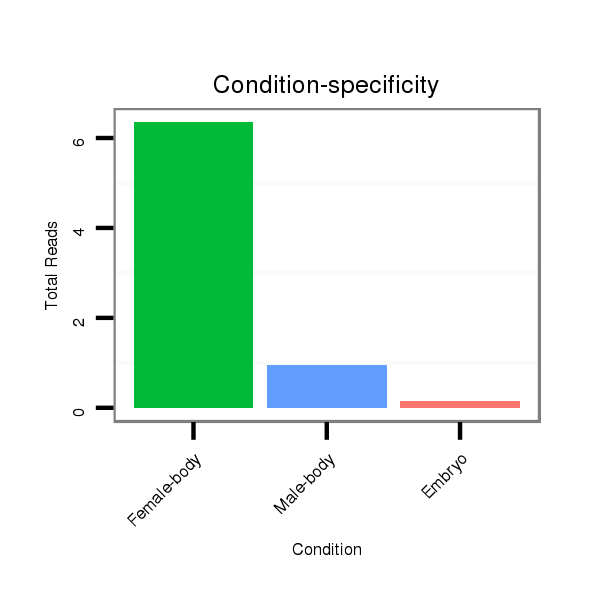
Read size | # Mismatch | Hit Count | Total Norm | Total | M042 female body |
V111 male body |
M021 embryo |
V042 embryo |
|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................CAACAGTTTTCCAGTGGAATCCGTC.......................................................................................... | 25 | 0 | 20 | 4.75 | 95 | 85 | 9 | 0 | 1 |
| ........................................................................................................................................CAACAGTTTTCCAGTGGAATCCGTCT......................................................................................... | 26 | 0 | 20 | 0.45 | 9 | 6 | 2 | 1 | 0 |
| ........................................................................................................................................CAACAGTTTTCCAGTGGAATCCGTCTA........................................................................................ | 27 | 0 | 20 | 0.40 | 8 | 8 | 0 | 0 | 0 |
| ........................................................................................................................................CAACAGTTTTCCAGTGGAATCCG............................................................................................ | 23 | 0 | 20 | 0.40 | 8 | 3 | 5 | 0 | 0 |
| ........................................................................................................................................CAACAGTTTTCCAGTGGAATCCGT........................................................................................... | 24 | 0 | 20 | 0.30 | 6 | 4 | 1 | 1 | 0 |
| .........................................................................................................................................................................TAAGACCTTCCTTAATCCGCTCGATG........................................................ | 26 | 0 | 20 | 0.25 | 5 | 5 | 0 | 0 | 0 |
| .........................................................................................................................................AACAGTTTTCCAGTGGAATCCGTC.......................................................................................... | 24 | 0 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 |
| .........................................................................................................................................................................TAAGACCTTCCTTAATCCGCTC............................................................ | 22 | 0 | 20 | 0.15 | 3 | 2 | 1 | 0 | 0 |
| .........................................................................................................................................................................TAAGACCTTCCTTAATCCGCTCGAT......................................................... | 25 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| ........................................................................................................................................CAACAGTTTTCCAGTGGAATCCGTCTAC....................................................................................... | 28 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| ..........................................................................................................................................ACAGTTTTCCAGTGGAATCCGTCTAC....................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................CAATTAAGACCTTCCTTAATCCGCTCGA.......................................................... | 28 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................CAACAGTTCTCCGGTGGAATCC............................................................................................. | 22 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................ACAGTTTTCCAGTGGAATCCGTC.......................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................TAATCCGCTCGATGTAGTTCCTTGCT............................................ | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................TAAGACCTTCCTTAATCCGCTCG........................................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................................................GACCTTCCTTAATCCGCTCGATGTAGT.................................................... | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................CAATTAAGACCTTCCTTAATCCGCTCG........................................................... | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................AATTAAGAGCTTCCTTAATCCGCTCGA.......................................................... | 27 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................GAACAGTTTTCCAGTGGAATCCGTC.......................................................................................... | 25 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................TAAGACCTTCCTTAATCCGCTCGATC........................................................ | 26 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................................CCTTAATCCGCTCGATGTAGTTCCT................................................ | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................CAACAGTTTTCCAGTGGAATCCGAC.......................................................................................... | 25 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................AACAGTTTTCCAGTGGAATCCGT........................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................CAACAGTTGTCCAGTGGAATCCGTCT......................................................................................... | 26 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................CTTCCTTAGTCCGCTCGATGTAG..................................................... | 23 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ................ACAACACGCCTTCTCGCGCT....................................................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................CAACAGTTATCCAGTGGAATCCGTC.......................................................................................... | 25 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................TAATCCGCTCGATGTAGTTCCTTGC............................................. | 25 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................CAACAGTTGTCCAGTGGAATCCGT........................................................................................... | 24 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................AATCCGCTCGATGTAGTTCCTTGC............................................. | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
|
ACGCAGACACGTAAGGTGTTGAGCGGAAGAACCCGAATGAGAATCGTGTCCACCCGAGGACTTTGTAAGAAATTTATACGCCATTCTCTCGAGTAGAATCTGTAAAAAGTATTTTGGACATGCTCGATTCAGGGAAGTTGTCAAAAGGTCACCTTAGGCAGATGGGTTAATTCTGGAAGGAATTAGGCGAGCTACATCAAGGAACGAGTAAGCAAAATAAAACATATAGAGAATTTAGATCACTAGTATTA
********************************************************.........((.((((((..............(((((......))))))))))).))..**************************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M042 female body |
V111 male body |
V042 embryo |
M021 embryo |
|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................AGTATTTTGACTATGCTCGA............................................................................................................................ | 20 | 3 | 19 | 0.95 | 18 | 7 | 2 | 6 | 3 |
| ...................................................................................................................................................................................GAATTAGGCGAGCTACATCAAGGAA............................................... | 25 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 |
| .................................................................................................................................................................................AGGAATTAGGCGAGCTACATCAAGGA................................................ | 26 | 0 | 20 | 0.15 | 3 | 1 | 2 | 0 | 0 |
| ............................................................................................................................CGATTCAGGGAAGTTGTCAAAA......................................................................................................... | 22 | 0 | 18 | 0.11 | 2 | 0 | 2 | 0 | 0 |
| ...................................................................................................................................................................................GAATTAGGCGAGCTACATCAAGGA................................................ | 24 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| .....................................................................................................................................................................................ATTAGGCGAGCTACATCAAGGAA............................................... | 23 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| ...............................CCCGAATGAGAATCGTGTCCAC...................................................................................................................................................................................................... | 22 | 0 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 |
| ...................TGAGCGGAAGAACCCGAATGAGAAT............................................................................................................................................................................................................... | 25 | 0 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................................................TTAGGCGAGCTACATCAAGGAA............................................... | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................GGTCACCTTAGGCAGATGGGT.................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................AACGAATTAGGCGAGGTACATCAA................................................... | 24 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droPer2 | scaffold_2131:497-747 + | dpe_1168 | TGCGTCTGTG-----------------------CATTCCACAACTCGCCTTCTTGGGCTTACTCTTAGCAC--------------AGGT----------GGGCTCCTGAAACATTCTTTAAATATGCGGTAAGAGAGCTCATCTTAGACATTTTTCATAAAACCTGT-----------ACGAGCTAAGTCCCTTCAACAGTTTTCCAGTGGAATCCGTCTACCCAATTAAGACCTTCCTTAATC-CGCTCGATGTAGTTCCTTGCTCATTCGTTTTATTTTGTATATCTCTTAAATCTAGTGATCATAAT |
| dp5 | 4_group3:4395820-4396067 - | GACACCTGGAGAAAACTAAAAAAAATATTGATATATTCCGTAGA-------------TTTATTTACAGTACTTTTTATGGAATAATAATAGAGTTATCTATTTTTATAATTAATTATTCAAATATTT----ATGAAGTTCAATTTAAATTATTTTGATCAAAATTATCGTATATATGCACATATTTTGTGGATTTATTTATAGTACAATGGAATAA-------TA-ATAAGAATATTATGAATTATAATTATTTATATTTTTAGTTAATTTTT------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
Generated: 06/08/2015 at 03:51 PM