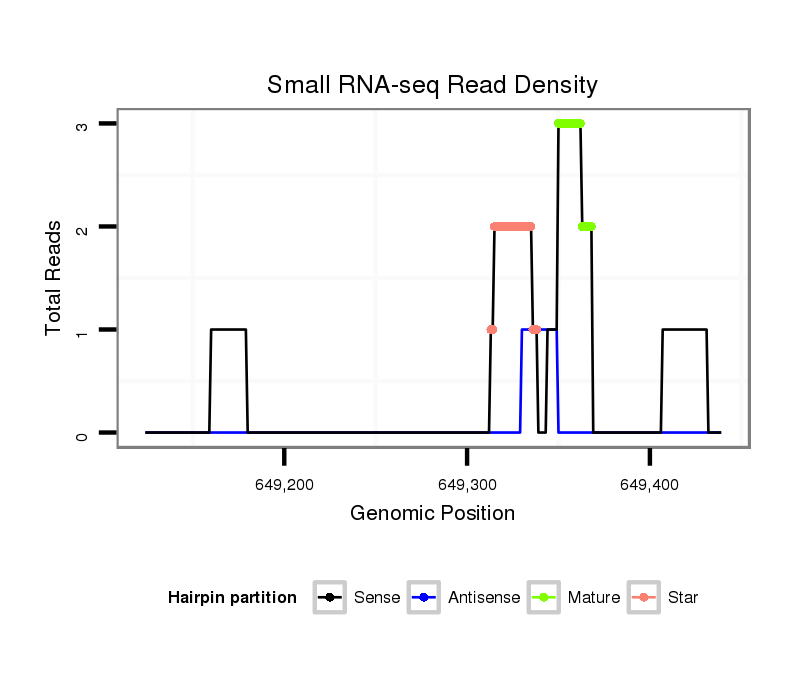
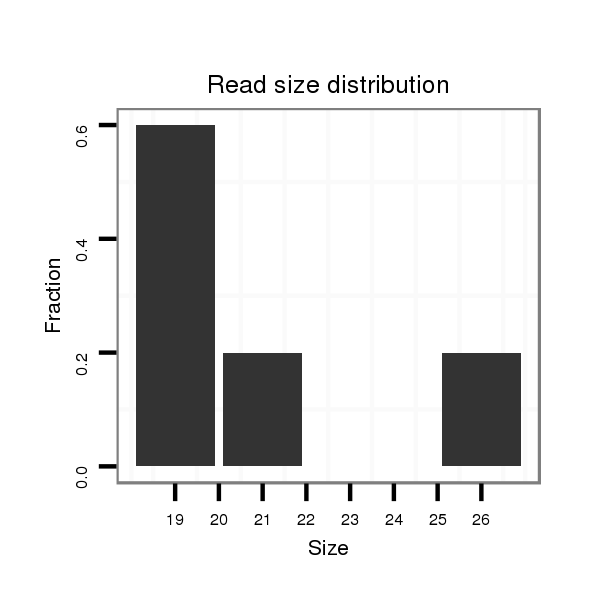
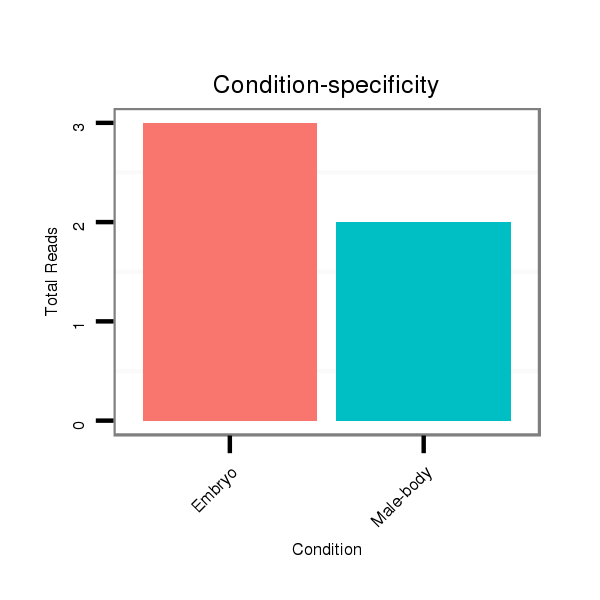
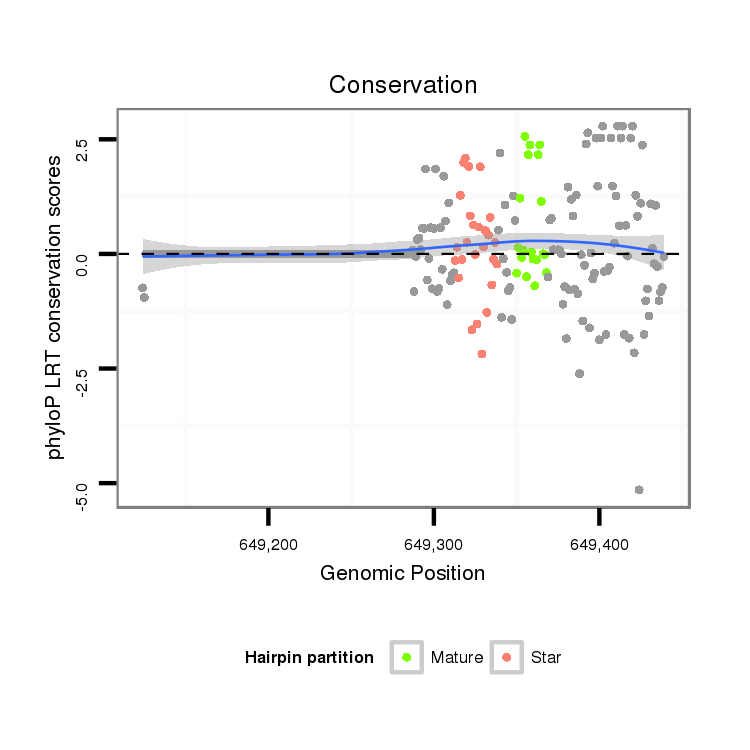
| G--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGGA---------T---ACT---TCTGATGA---CGATGA--------------TGATGATGATG------------------ATG---------------ACG---------------------ACGACGA---------------------------------CGATGATGATGAT---------------------------GATGATGATGACGA---CGACGATGATG---ACGACGAAGAGGAGGACGATGATGTGGAAG--------------TGGC | Size | Hit Count | Total Norm | Total | M020
Head | M045
Female-body | V117
Male-body | V118
Embryo | V119
Head |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................CGAAGAGGAGGACGATGATGTGG..................... | 23 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................GACGATGATG---ACGACGAAGAGGAGGAC............................... | 27 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................GATGACGA---CGACGATGATG---ACGACG.......................................... | 25 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................................................................................................................GGACGATGATGTGGAAG--------------TGGC | 21 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................ACGATGATG---ACGACGAAGA...................................... | 19 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................GATGAT---------------------------GATGATGATGACGA---CGACGAT....................................................... | 27 | 6 | 0.67 | 4 | 0 | 0 | 4 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................GATGATG---ACGACGAAGAGGA................................... | 20 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................ATGATG---ACGACGAAGAGG.................................... | 18 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................GAT---------------------------GATGATGATGACGA---CGACGAT....................................................... | 24 | 7 | 0.29 | 2 | 0 | 0 | 1 | 1 | 0 |
| ..............................................................................................................................................................................................................................................................................................G---------------------ACGACGA---------------------------------CGATGATGATGAT---------------------------............................................................................... | 21 | 20 | 0.25 | 5 | 0 | 0 | 5 | 0 | 0 |
| ..............................................................................................................................................................................................................................................GATGATGATG------------------ATG---------------ACG---------------------ACGAC.......................................................................................................................................................... | 21 | 20 | 0.20 | 4 | 2 | 0 | 2 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................GAT---------------------------GATGATGATGACGA---CGAC.......................................................... | 21 | 20 | 0.20 | 4 | 2 | 0 | 2 | 0 | 0 |
| .................................................................................................................................................................................................................................................GATGATG------------------ATG---------------ACG---------------------ACGACGA---------------------------------CGAT................................................................................................................... | 24 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................................................CTGATGA---CGATGA--------------TGATGATGATG------------------A........................................................................................................................................................................................................ | 25 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................GATGATGACGA---CGACGATGA..................................................... | 20 | 19 | 0.11 | 2 | 0 | 0 | 2 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................GATGATGATGACGA---CGACGA........................................................ | 20 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 |
| ....................................................................................................................................................................................................................GA---CGATGA--------------TGATGATGATG------------------ATG---------------ACG---------------------............................................................................................................................................................... | 25 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................T---------------------------GATGATGATGACGA---CGACGAT....................................................... | 22 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................GATGAT---------------------------GATGATGATGACGA---CGA........................................................... | 23 | 20 | 0.10 | 2 | 0 | 1 | 0 | 1 | 0 |
| .................................................................................................................................................................................................................................................GATGATG------------------ATG---------------ACG---------------------ACGAC.......................................................................................................................................................... | 18 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................GATGATGATGACGA---CGAC.......................................................... | 18 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................GATGATG------------------ATG---------------ACG---------------------ACGACGA---------------------------------....................................................................................................................... | 20 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................CGA---------------------------------CGATGATGATGAT---------------------------GATGATG........................................................................ | 23 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................GATGATGACGA---CGACGATGATG---................................................ | 22 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................................................................................GA---CGACGATGATG---ACGACG.......................................... | 19 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................................................................................................GATG------------------ATG---------------ACG---------------------ACGACGA---------------------------------CGATG.................................................................................................................. | 22 | 14 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................GATGATGATGACGA---CGACGAT....................................................... | 21 | 15 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................................GATGATGAT---------------------------GATGATGATGACGA---C............................................................. | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................................................TGATGATG------------------ATG---------------ACG---------------------ACGAC.......................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................................................................................................GATGATGATG------------------ATG---------------ACG---------------------ACGACGA---------------------------------CG..................................................................................................................... | 25 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................................................................................................................ATG---------------ACG---------------------ACGACGA---------------------------------CGATGATG............................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................................................................................ATGATGATG------------------ATG---------------ACG---------------------ACGAC.......................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................GAT---------------------------GATGATGATGACGA---CGA........................................................... | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................................................GA--------------TGATGATGATG------------------ATG---------------ACG---------------------ACGA........................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................ATGATGACGA---CGACGATGA..................................................... | 19 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................GATGATGACGA---CGACGAT....................................................... | 18 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................A---------------------------------CGATGATGATGAT---------------------------GATGAT......................................................................... | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................................GATGATGAT---------------------------GATGATGATGAC................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................GATGA--------------TGATGATGATG------------------ATG---------------ACG---------------------AC............................................................................................................................................................. | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................................................................GATGA--------------TGATGATGATG------------------ATG---------------AC..................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................T---------------------------GATGATGATGACGA---CGAC.......................................................... | 19 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................TGATGAT---------------------------GATGATGATGACG.................................................................. | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................................................................................................................GATGATGATG------------------ATG---------------ACG---------------------ACGA........................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................................................................................................................................TG---------------ACG---------------------ACGACGA---------------------------------CGATGA................................................................................................................. | 18 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................................................GA--------------TGATGATGATG------------------ATG---------------ACG---------------------AC............................................................................................................................................................. | 21 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................................................................................GA--------------TGATGATGATG------------------ATG---------------ACG---------------------A.............................................................................................................................................................. | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................A---------------------------------CGATGATGATGAT---------------------------GATGATGAT...................................................................... | 23 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................................GA---CGATGA--------------TGATGATGAT............................................................................................................................................................................................................................ | 18 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................ATGA--------------TGATGATGATG------------------ATG---------------AC..................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................................................................................................................GATGATGATG------------------ATG---------------ACG---------------------ACG............................................................................................................................................................ | 19 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................CGACGA---------------------------------CGATGATGATGAT---------------------------G.............................................................................. | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................................................................................................................................................................ACGACGA---------------------------------CGATGATGATGAT---------------------------............................................................................... | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................GATGAT---------------------------GATGATGATGACGA---C............................................................. | 21 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................CGACGA---------------------------------CGATGATGATGAT---------------------------GATGA.......................................................................... | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................................................................................................................CG---------------------ACGACGA---------------------------------CGATGATGATGAT---------------------------GATG........................................................................... | 26 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................................................................................................................................TG---------------ACG---------------------ACGACGA---------------------------------CGATGAT................................................................................................................ | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................GAT---------------------------GATGATGATGACGA---CG............................................................ | 19 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |