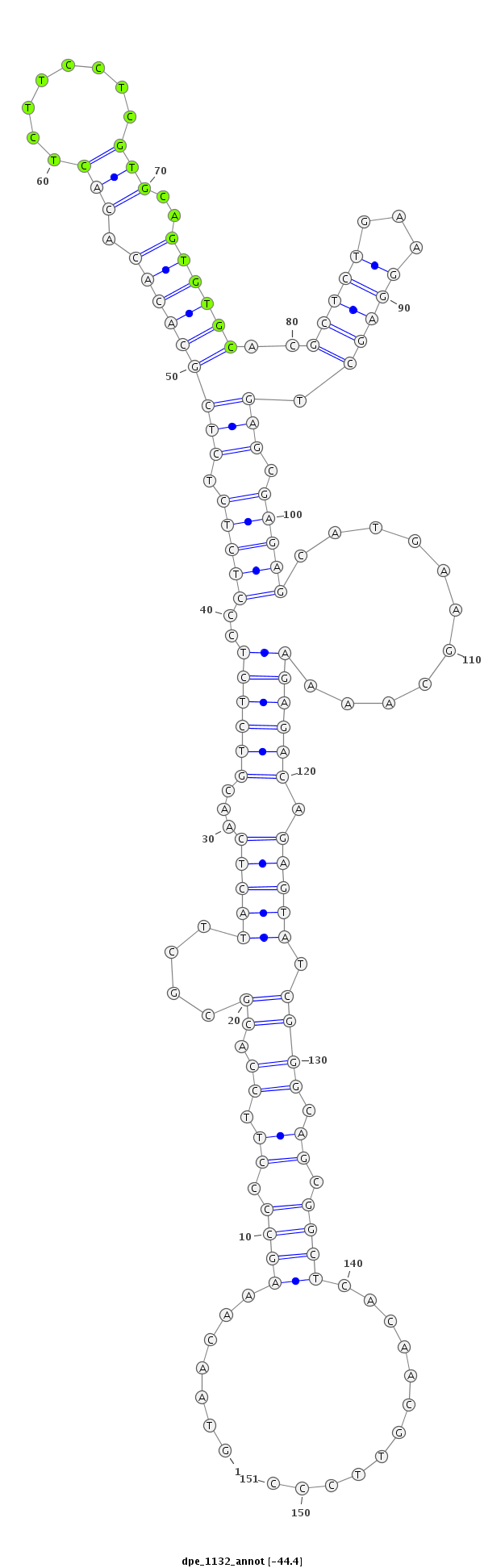
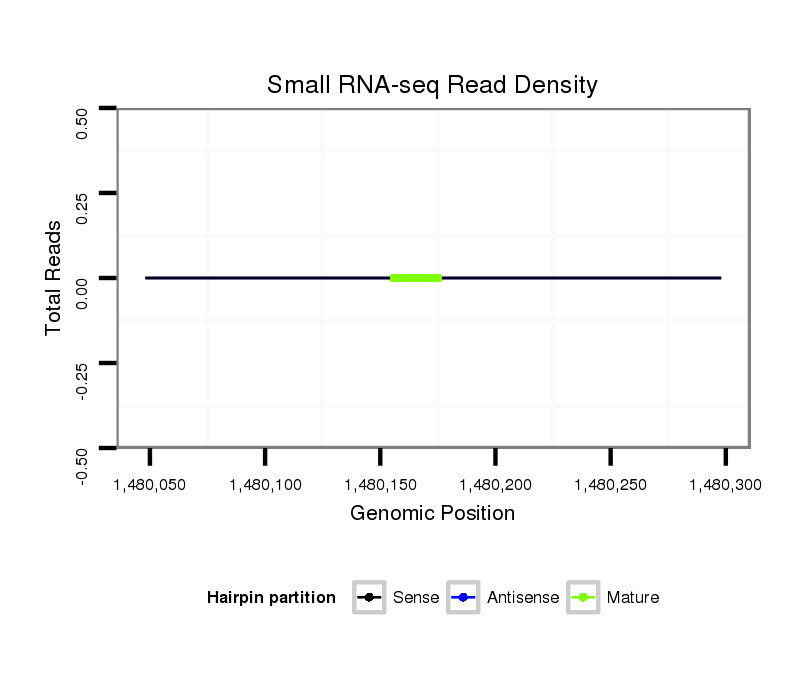
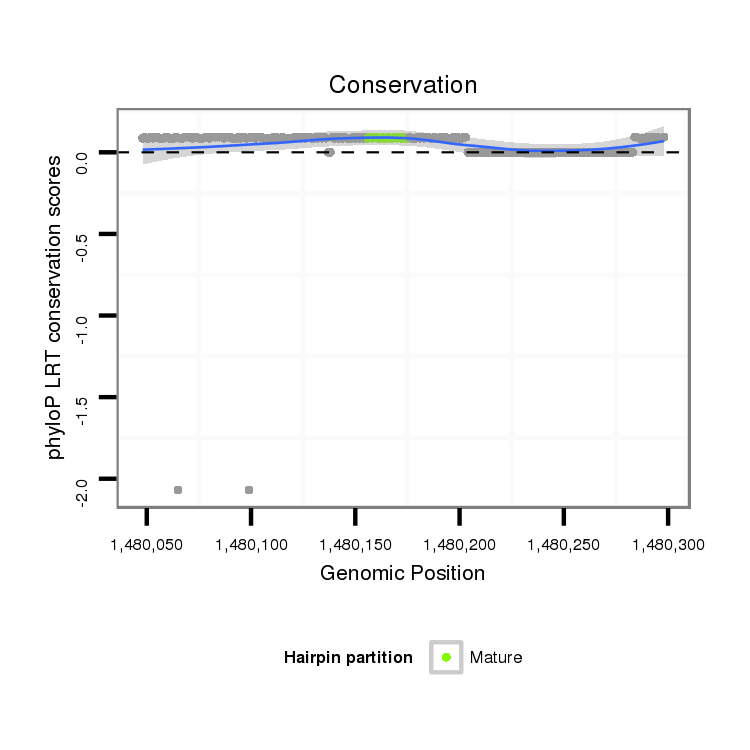
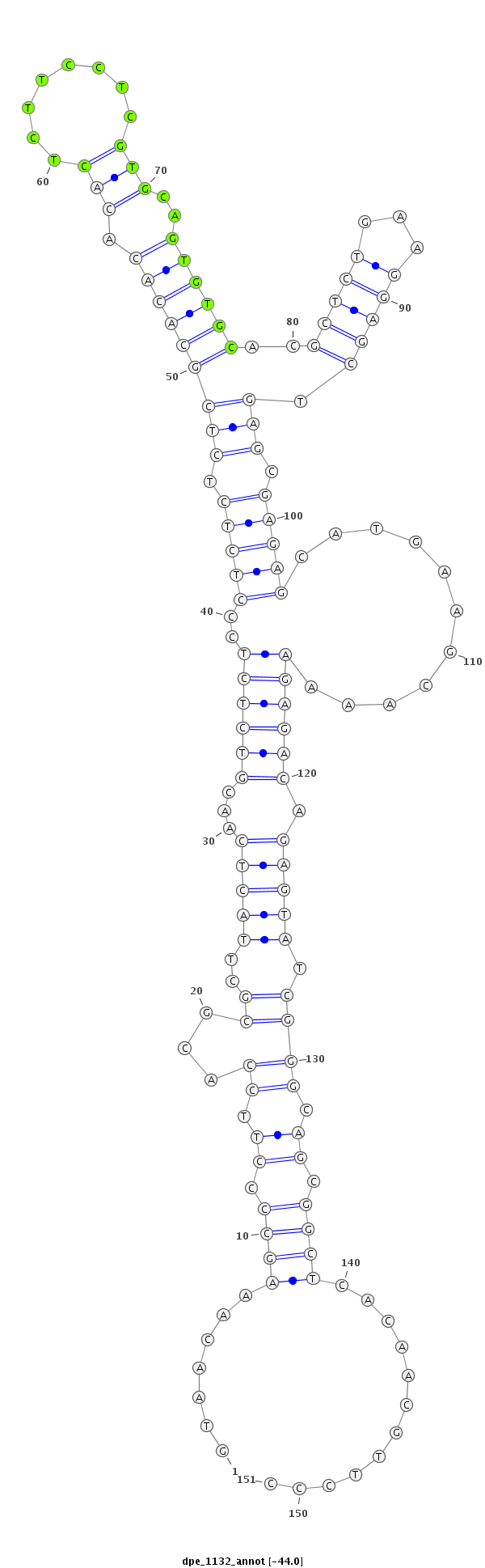
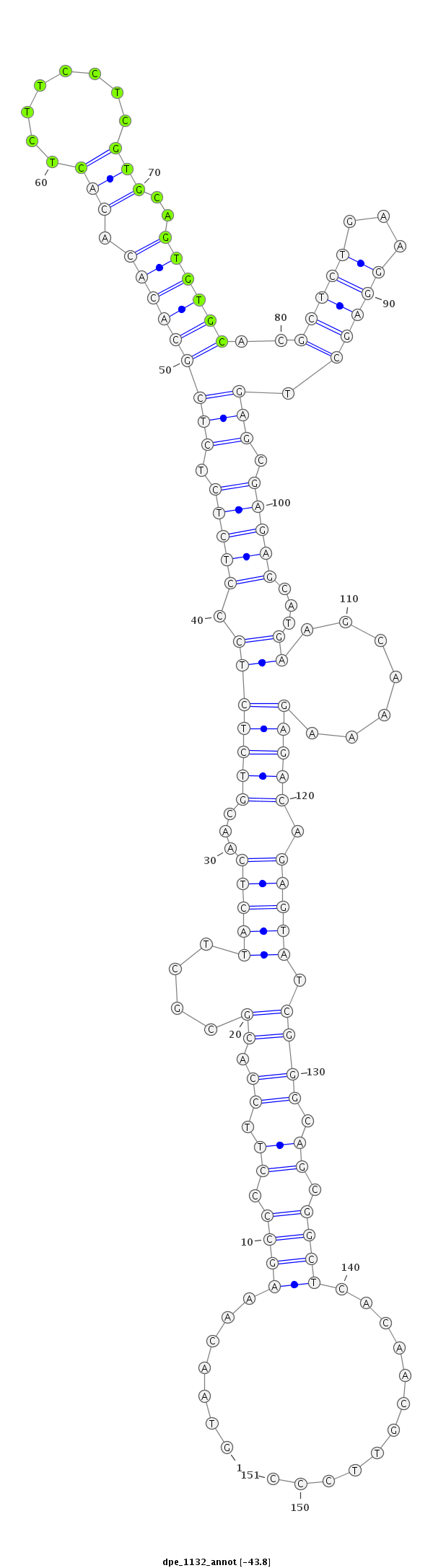
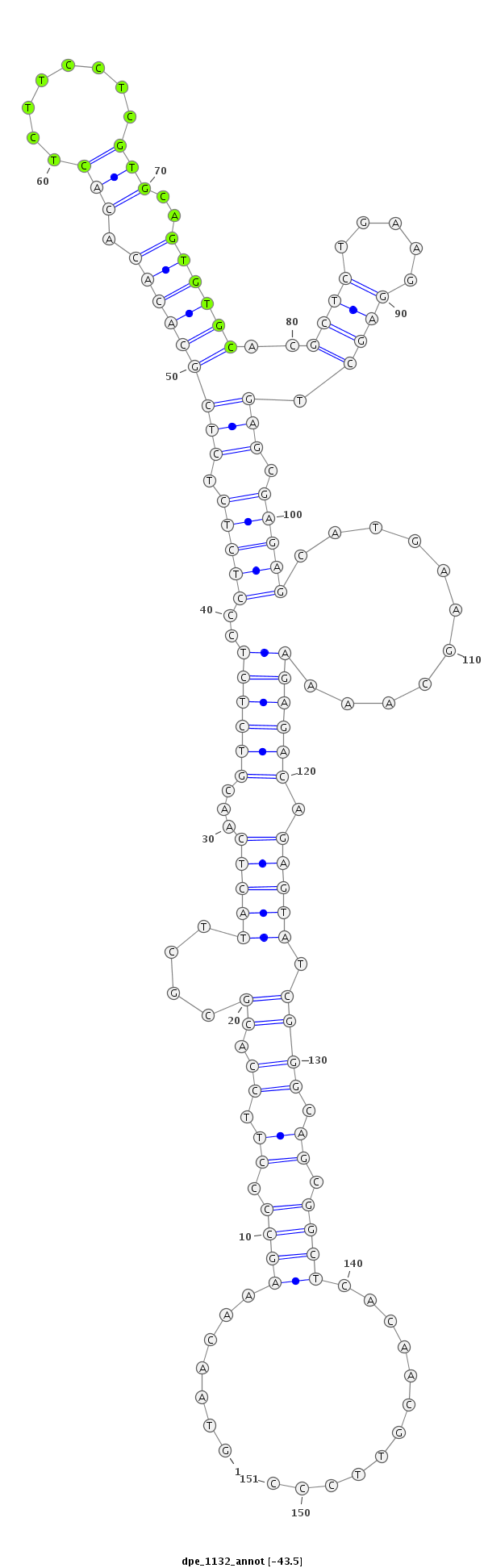
ID:dpe_1132 |
Coordinate:scaffold_20:1480098-1480248 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -44.0 | -43.8 | -43.5 |
 |
 |
 |
CDS [Dper\GL22891-cds]; exon [dper_GLEANR_5137:1]; intron [Dper\GL22891-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GACGAGAAGGGTAGCCATGGATTAGATCGGATCTTTATTGTAAGCCAGAAGTAACAAAGCCCCTTCCACGCGCTTACTCAACGTCTCTCCCTCTCTCTCGCACACACACTCTTCCTCGTGCAGTGTGCACGCTCTGAAGGAGCTGAGCGAGAGCATGAAGCAAAAGAGACAGAGTATCGGGCAGCGGCTCACAACGTTCCCGCTCGTTTTACACTCTATTTACATATGCCCTTGTTAACGGCTCTAGTGCA **************************************************.......((((.((.((.((....(((((...((((((..(((((.(((((((((.(((........)))..))))))..(((((...))))).))).)))))...........)))))).))))).)))).)).))))............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V057 head |
V042 embryo |
|---|---|---|---|---|---|---|---|
| ...................................................TATCAAAGCCCCTTTCACGCGTT................................................................................................................................................................................. | 23 | 3 | 1 | 1.00 | 1 | 1 | 0 |
| ............................................................................................................CTCTTCCTCGTGCAGTGTGC........................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 1 |
|
CTGCTCTTCCCATCGGTACCTAATCTAGCCTAGAAATAACATTCGGTCTTCATTGTTTCGGGGAAGGTGCGCGAATGAGTTGCAGAGAGGGAGAGAGAGCGTGTGTGTGAGAAGGAGCACGTCACACGTGCGAGACTTCCTCGACTCGCTCTCGTACTTCGTTTTCTCTGTCTCATAGCCCGTCGCCGAGTGTTGCAAGGGCGAGCAAAATGTGAGATAAATGTATACGGGAACAATTGCCGAGATCACGT
**************************************************.......((((.((.((.((....(((((...((((((..(((((.(((((((((.(((........)))..))))))..(((((...))))).))).)))))...........)))))).))))).)))).)).))))............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V057 head |
V111 male body |
V050 head |
V042 embryo |
M042 female body |
M021 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................TTTCGGCGAGAGTGCGCGAATG.............................................................................................................................................................................. | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................CGTCATTGTGTCGGGGAAGGC....................................................................................................................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................AGAGGGAGAGAGAGCGTG.................................................................................................................................................... | 18 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................GTACGCGGGAACAATTGA........... | 18 | 3 | 20 | 0.30 | 6 | 1 | 0 | 0 | 0 | 3 | 2 |
| ...................................................................................................................................................ACACTCGTACTTCGTTTTC..................................................................................... | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................TCGTTATTGTGTCGGGGAAGGT....................................................................................................................................................................................... | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................TGGCCGATTGTTGCGAGGGC................................................. | 20 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................TTTCGGGGAAGGTGTGCGAGTGT............................................................................................................................................................................. | 23 | 3 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................ACCCTCGTACTTCGTTTCCT.................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................GGCAAACGTGCGAGAGTTC................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................GGTACGCGGGAACAATTG............ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droPer2 | scaffold_20:1480048-1480298 + | dpe_1132 | GACGAGAAGGGTAGCCATGGATTAGATCGGATCTTTATTGTAAGCCAGAAGTAACAAAGCCCCTTCCACGCGCTTACTCAACGTCTCTCCCTCTCTCTCGCACACACACTCTTCCTCGTGCAGTGTGCACGCTCTGAAGGAGCTGAGCGAGAGCATGAAGCAAAAGAGACAGAGTATCGGGCAGCGGCTCACAACGTTCCCGCTCGTTTTACACTCTATTTACATATGCCCTTGTTAACGGCTCTAGTGCA |
| dp5 | XR_group6:11580646-11580814 + | GACGAGAAGGGTAGCCAGGGATTAGATCGGATCTTTATTGTAAGCCAGAAGGAACAAAGCCCCTTCCACGCGCTTACTCAACGTCTCTC--TCTCTCTCGCACACACACTCTTCCTCGTGCAGTGTGCACGCTCTGAAGGAGCTGAGCGAGAGCAT--------------------------------------------------------------------------------AACGGCTCTAGTGCA |
| Species | Read alignment | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
||||||||||||||||||||||
| dp5 |
|
Generated: 06/08/2015 at 03:47 PM