
ID:dpe_1098 |
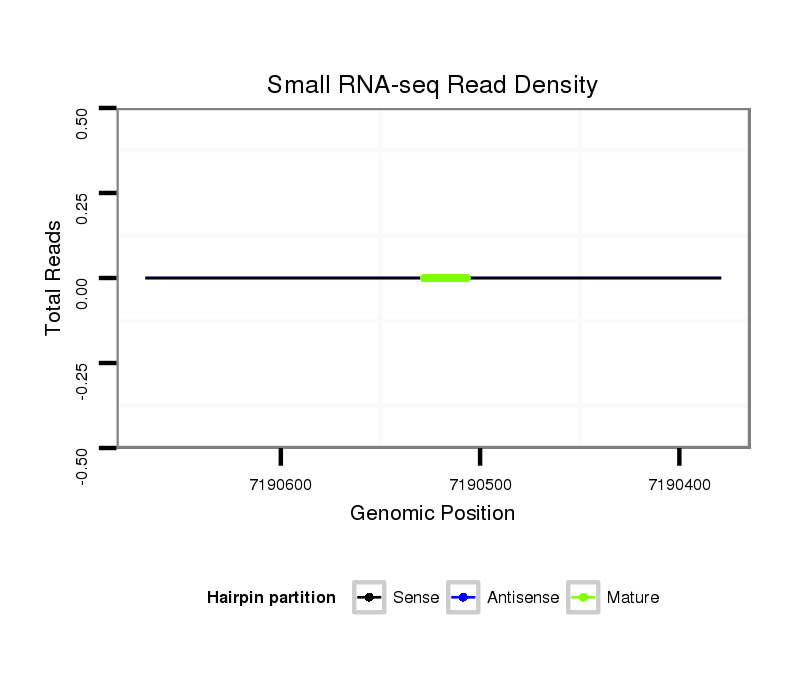
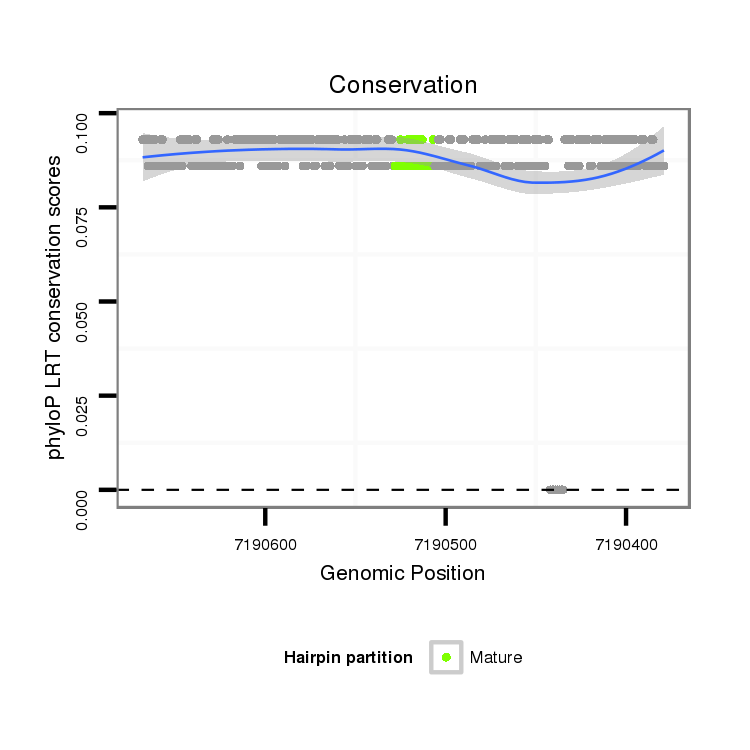
Coordinate:scaffold_2:7190429-7190618 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -34.8 | -34.7 | -34.7 |
 |
 |
 |
exon [dper_GLEANR_10608:1]; CDS [Dper\GL10481-cds]; exon [dper_GLEANR_10608:2]; CDS [Dper\GL10481-cds]; intron [Dper\GL10481-in]
No Repeatable elements found
| mature | star |
| -----------------------------#####################----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## AAAGTACAGGAACCGCCGGGGAGTTTACGATGGCCGCCGATCAGGCGAGGGTATCAAAAAATAAAAGAAGAAGACCTAAGAAAAAAATAGTCAATGCAAAAGATGAATAGGGAGGAAACAAGGAAAACGTACCCGGTTTGGGCTGGCTCATGATGACGCGGTGGTAGCCGAACGCCAGAGCCGAATACCCTGGAGTCTAATCCAGAATCCATGTACGAACTGGCATATCTATATATGTAGCTCTATAAGCTAATCGTGATCTACAAGACTCAGAGCTATCGCAACGCGGG **************************************************............................................((((..(((((..(((.......(((.(((....(((..((((((.((((((((.((.((......)).)))))))...))).))))))..))).(((((......)))))..))).))).....)))...))))).....)))).************************************************** |
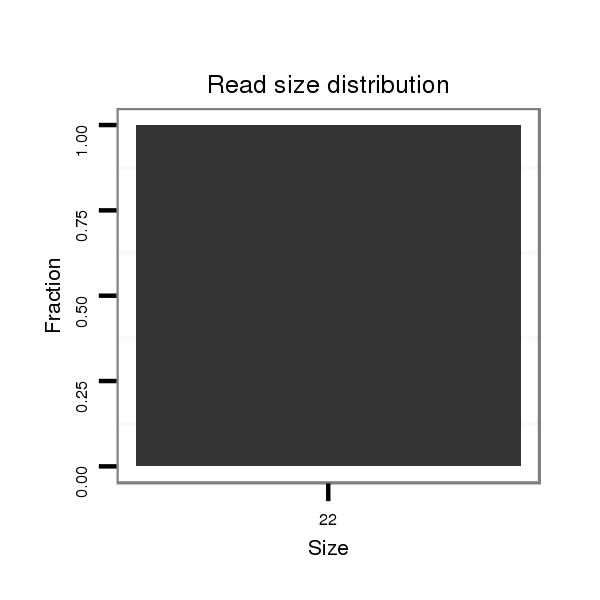
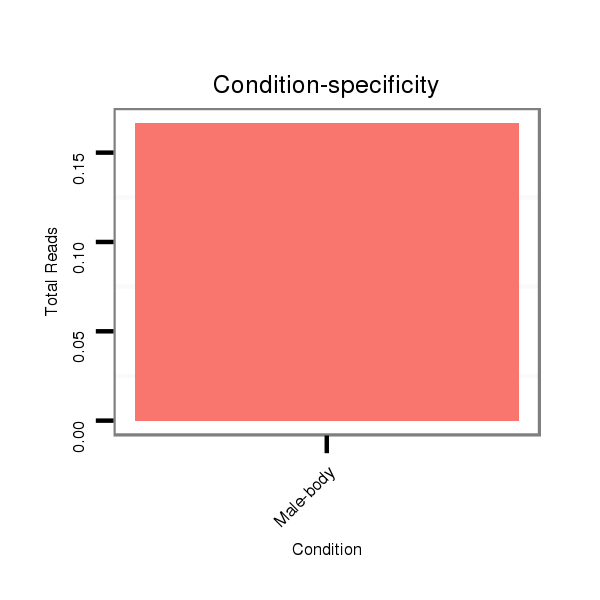
Read size | # Mismatch | Hit Count | Total Norm | Total | M042 female body |
V111 male body |
V057 head |
M021 embryo |
|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................................TCTAAGTCAGAATCCATG............................................................................. | 18 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................CTCTAAGTCAGAATCCATG............................................................................. | 19 | 3 | 19 | 0.26 | 5 | 2 | 1 | 0 | 2 |
| ............................................................................................................................................GGCTGGCTCATGATGACGCGGT................................................................................................................................ | 22 | 0 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................................AATACCCGGGAGTATAAGC........................................................................................ | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................................................................................................................................TACAACACTCAGAAATATCGC........ | 21 | 3 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 |
|
TTTCATGTCCTTGGCGGCCCCTCAAATGCTACCGGCGGCTAGTCCGCTCCCATAGTTTTTTATTTTCTTCTTCTGGATTCTTTTTTTATCAGTTACGTTTTCTACTTATCCCTCCTTTGTTCCTTTTGCATGGGCCAAACCCGACCGAGTACTACTGCGCCACCATCGGCTTGCGGTCTCGGCTTATGGGACCTCAGATTAGGTCTTAGGTACATGCTTGACCGTATAGATATATACATCGAGATATTCGATTAGCACTAGATGTTCTGAGTCTCGATAGCGTTGCGCCC
**************************************************............................................((((..(((((..(((.......(((.(((....(((..((((((.((((((((.((.((......)).)))))))...))).))))))..))).(((((......)))))..))).))).....)))...))))).....)))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droPer2 | scaffold_2:7190379-7190668 - | dpe_1098 | AAAGTACAGGAACCGCCGGGGAGTTTACGATGGCCGCCGATCAGGCGAGGGTATCAAAAAATAAAAGAAGAAGACCTAAGAAAAAAATAGTCAATGCAAAAGATGAATAGGGAGGAAACAAGGAAAACGTACCCGGTTTGGGCTGGCTCATGATGACGCGGTGGTAGCCGAACGCCAGAGCCGAATACCCTGGAGTCTAATCCAGAATCCATGTACGAACTGGCATATCTATATATGTAGCTCTATAAGCTAATCGTGATCTACAAGACTCAGAGCTATCGCAACGCGGG |
| dp5 | 3:6989224-6989505 - | dps_1065 | AAAGTACAGGAACCGCCGGGGAGTTTACGATGGCCGCCGATCAGGCGAGGGTATCAAAAAATAAAAGAAGAAGACCTAAGAAAAAAATAGTCAATGCAAAAGATGAATAGGGAGGAAACAAGGAAAACGTACCCGGTTTGGGCTGGCTCATGATGACGCGGTGGTAGCCGAACGCCAGAGCCGAATACCCTGGAGTCTAATCCAGAATCCATGTACGAACTGGCAT--------ATGTAGCTCTATAAGCTAATCGTGATCTACAAGACTCAGAGCTATCGCAACGCGGG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
Generated: 06/08/2015 at 03:46 PM