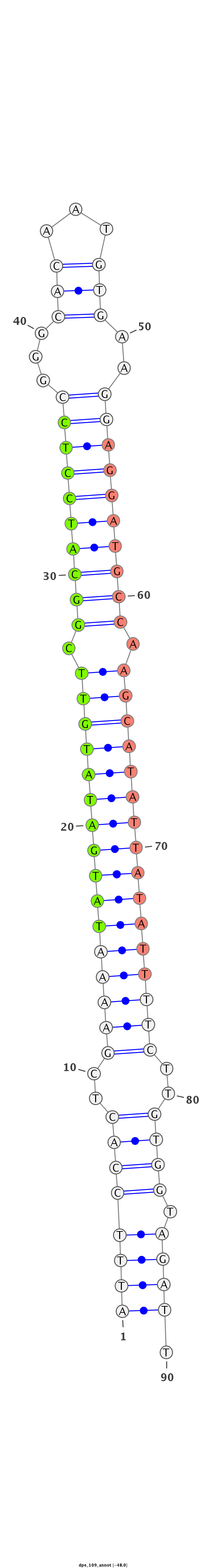

ID:dpe_109 |
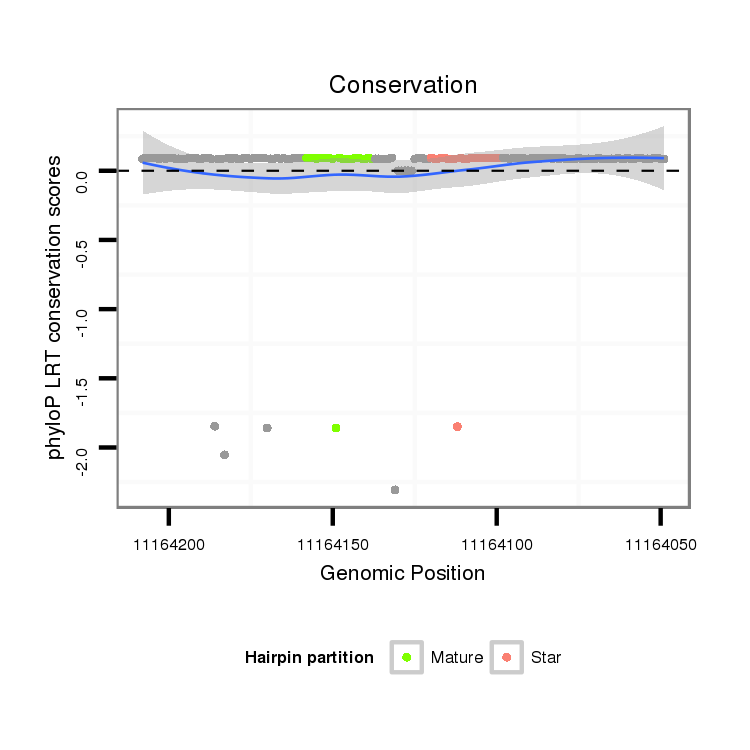
Coordinate:scaffold_0:11164099-11164158 - |
Confidence:Known Ortholog |
Class:Canonical miRNA |
Genomic Locale:intergenic |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
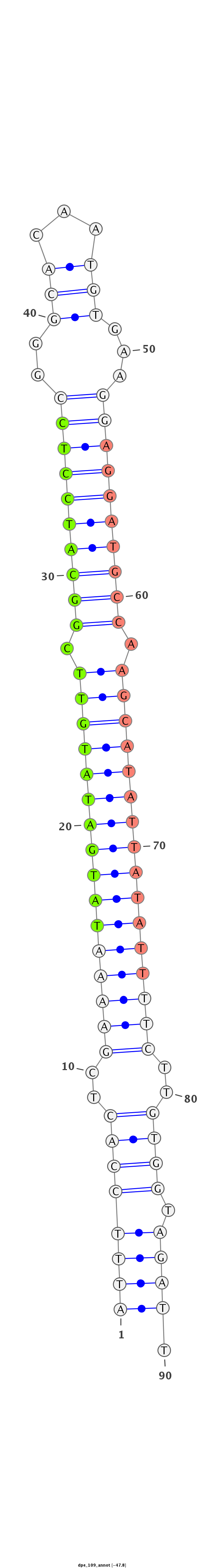
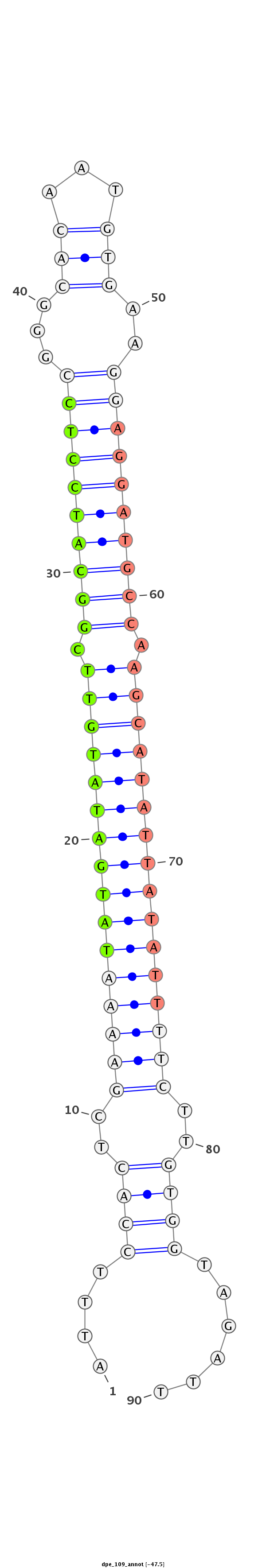
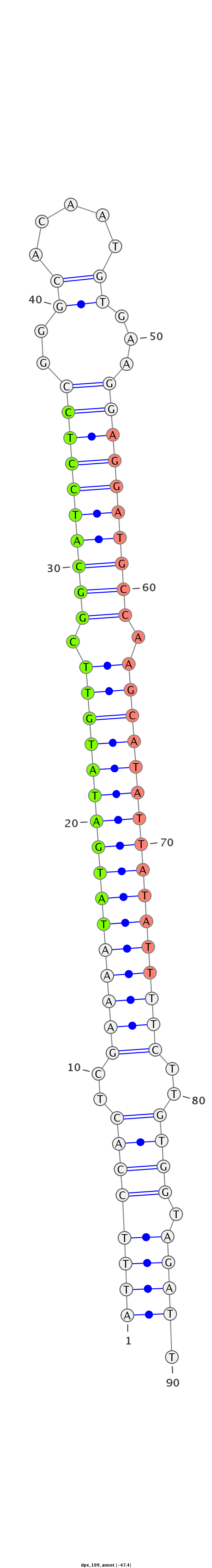
| -47.8 | -47.5 | -47.4 |
 |
 |
 |
intergenic
No Repeatable elements found
|
CTTATCAGATCCGCAATGCTTCCCGATCACCATGAATTTCCACTCGAAAATATGATATGTTCGGCATCCTCCGGGCACAATGTGAAGGAGGATGCCAAGCATATTATATTTTCTTGTGGTAGATTCTCAGCGCAGCGCCACCGGCTGGAGGAAGCCATGG
***********************************((((((((..((((((((((((((((.((((((((((...(((...)))..)))))))))).))))))))))))))))..)))).)))).*********************************** |
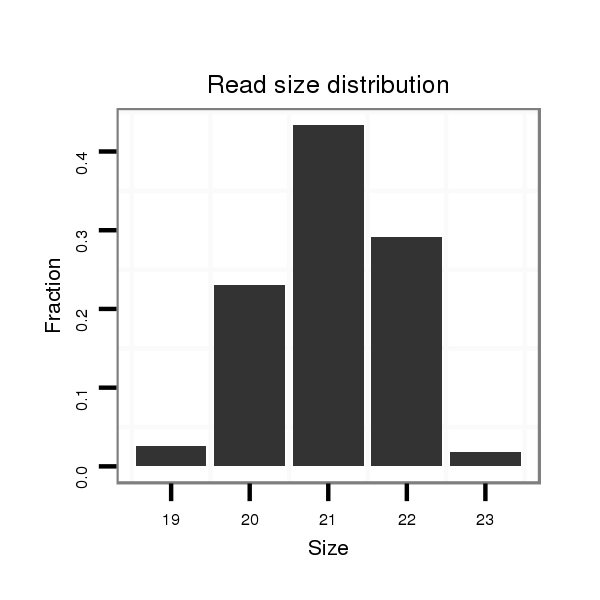
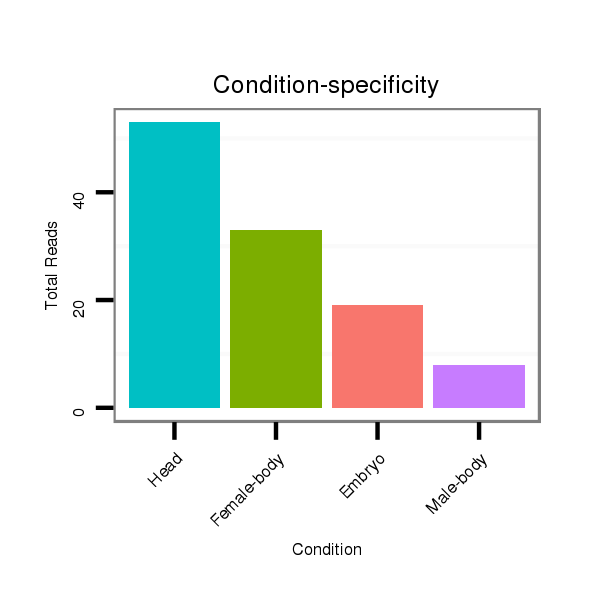
Read size | # Mismatch | Hit Count | Total Norm | Total | V057 head |
M042 female body |
M021 embryo |
V050 head |
V111 male body |
|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................TATGATATGTTCGGCATCCTC......................................................................................... | 21 | 0 | 1 | 31.00 | 31 | 8 | 6 | 15 | 2 | 0 |
| ..................................................TATGATATGTTCGGCATCCTCC........................................................................................ | 22 | 0 | 1 | 31.00 | 31 | 10 | 9 | 1 | 6 | 5 |
| ...................................................ATGATATGTTCGGCATCCTC......................................................................................... | 20 | 0 | 1 | 18.00 | 18 | 12 | 3 | 1 | 2 | 0 |
| ...................................................ATGATATGTTCGGCATCCTCC........................................................................................ | 21 | 0 | 1 | 16.00 | 16 | 5 | 8 | 0 | 3 | 0 |
| ..................................................TATGATATGTTCGGCATCCT.......................................................................................... | 20 | 0 | 1 | 6.00 | 6 | 1 | 1 | 2 | 1 | 1 |
| .................................................ATATGATATGTTCGGCATCCT.......................................................................................... | 21 | 0 | 1 | 3.00 | 3 | 1 | 1 | 0 | 1 | 0 |
| ...................................................GTGATATGTTCGGCATCCTC......................................................................................... | 20 | 1 | 1 | 3.00 | 3 | 2 | 0 | 0 | 0 | 1 |
| ..................................................TATGATATGTTCGGCATCC........................................................................................... | 19 | 0 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 | 0 |
| ..................................................TGTGATATGTTCGGCATCCTCC........................................................................................ | 22 | 1 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 | 0 |
| ........................................................................................AGGATGCCAAGCATATTATATT.................................................. | 22 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 |
| ........................................................................................AGGATGCCAAGCATATTATAT................................................... | 21 | 0 | 1 | 2.00 | 2 | 0 | 1 | 0 | 1 | 0 |
| ..................................................TATGATATGTTAGGCATCCTC......................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TATGATATGTTCTACATCCT.......................................................................................... | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................TATGATATGTTCGGCATCCTCA........................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TATGATATGTTCGGCATCCTCCT....................................................................................... | 23 | 1 | 3 | 1.00 | 3 | 3 | 0 | 0 | 0 | 0 |
| ..................................................TATGATATGTTCGGCATCCTCCA....................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................AGGATGCCAAGCATATTATA.................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TATGATATGTTGGGCATCTTC......................................................................................... | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TATGGTATGTTCGGCATCCTC......................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TATGAGATGTTCGGCATCCT.......................................................................................... | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TATGATATATTCGGCATCCTC......................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................ATATGATATGTTCGGCATCCTC......................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................TGAAGGAGGATGCCAAGCAT.......................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..................................................TATGATATGTTCTGCATCCTCC........................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................CAATGTGAAGGAGGATGCCAAGC............................................................ | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 |
| ...................................................ATGATATGTTCGGCATCCTCCA....................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TATGATATGTTCGGTATCCTCC........................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................GTGATATGTTCGGCATCCTCC........................................................................................ | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................ACAATGTGAAGGAGGATGCCAAG............................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................ATGATATGGTCGGCATCCTC......................................................................................... | 20 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TATGAGATGGTCGGCATCGTC......................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................CCACTAGGAAATATGATAT...................................................................................................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................ACAATGCCAAGTAGGATGCC................................................................ | 20 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 |
| .......................................CCACTAGGAAATATGATATC..................................................................................................... | 20 | 3 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 |
|
GAATAGTCTAGGCGTTACGAAGGGCTAGTGGTACTTAAAGGTGAGCTTTTATACTATACAAGCCGTAGGAGGCCCGTGTTACACTTCCTCCTACGGTTCGTATAATATAAAAGAACACCATCTAAGAGTCGCGTCGCGGTGGCCGACCTCCTTCGGTACC
***********************************((((((((..((((((((((((((((.((((((((((...(((...)))..)))))))))).))))))))))))))))..)))).)))).*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M042 female body |
V057 head |
V111 male body |
V042 embryo |
|---|---|---|---|---|---|---|---|---|---|
| .......................................GGTGATCCTTTATACTATA...................................................................................................... | 19 | 2 | 3 | 5.33 | 16 | 16 | 0 | 0 | 0 |
| ..........................AGTGGTACTTGAAGGTGAGC.................................................................................................................. | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ..........................................................TTTGCCGTAGGAGGCCCGTGTT................................................................................ | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ....................................AAGGGTGATCCTTTATACTA........................................................................................................ | 20 | 3 | 9 | 1.00 | 9 | 8 | 0 | 1 | 0 |
| ......TCTAGGCGTTACGAACGACAAG.................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| GAATAGTCTAGGCGTTACGAA........................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................AAGGGTGATCCTTTATACTAT....................................................................................................... | 21 | 3 | 4 | 0.75 | 3 | 3 | 0 | 0 | 0 |
| .......................................GGTGATCCTTTATACTATAG..................................................................................................... | 20 | 3 | 13 | 0.69 | 9 | 9 | 0 | 0 | 0 |
| ............................................................................................................................ACATTCGCGTCGCGGTGCCC................ | 20 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................................................AAATCGCGTCGCGGTCGCCGACC............ | 23 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 |
| ...TAGTCTAGAGGTTACGAA........................................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 |
| .......................................GGTGATCCTTTATACTAT....................................................................................................... | 18 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................GTCGCGGGGGCCCACCTGCT........ | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droPer2 | scaffold_0:11164049-11164208 - | dpe_109 | confident | CTTATCAGATCCGCAATGCTTCCCGATCACCATGAATTTCCACTCGAAAATATGATATGTTCGGCATCCTCCGGGCACAATGTGAAGGAGGATGCCAAGCATATTATATTTTCTTGTGGTAGATTCTCAGCGCAGCGCCACCGGCTGGAGGAAGCCATGG |
| dp5 | 2:17318651-17318805 - | dps-mir-2503 | Known_mirbase | CTTATCAGATCCGCAATGCTTCTCGCTCACCATGAATTCCCACTCGAAAATATGATATGCTCGGCATCCTCCGGGCAG-----GAAGGAGGATGCCGAGCATATTATATTTTCTTGTGGTAGATTCTCAGCGCAGCGCCACCGGCTGGAGGAAGCCATGG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
Generated: 10/20/2015 at 06:45 PM