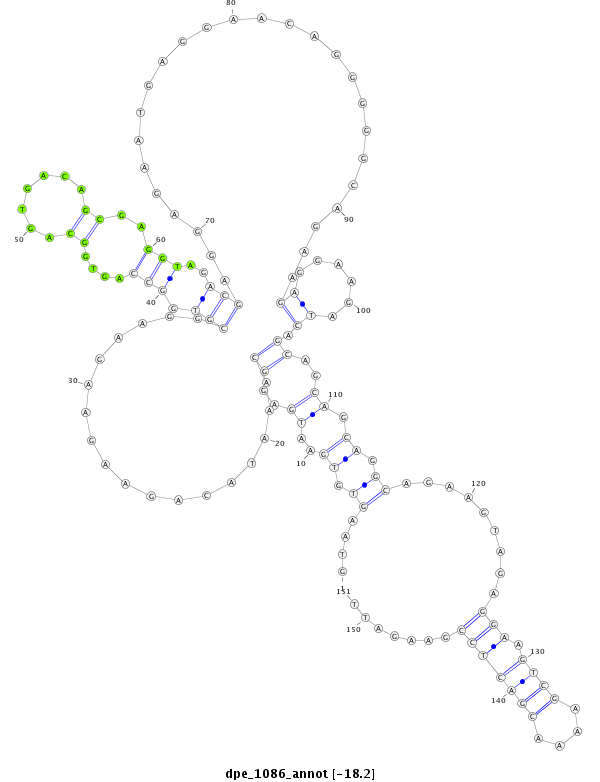
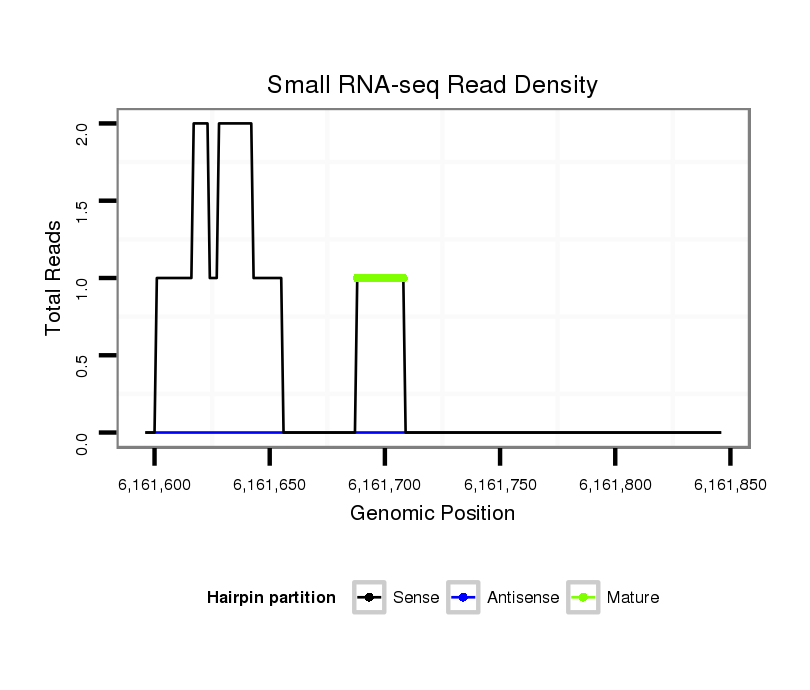
| droPer2 |
scaffold_2:6161596-6161846 + |
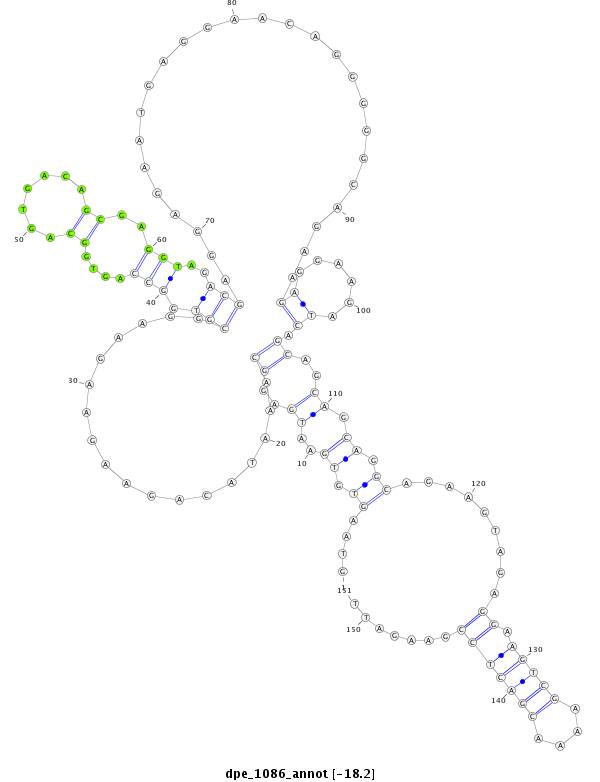
dpe_1086 |
ATCAGG------------------------CGTCATCTCCAGTGCGTAC---AGCGACAGCAGCGACGCGAGCCGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTG---GCAGTGACAGCGAGGTAGACGAGGAGAA---T---------------------GAGGAACAGGGGGCA------GAAGA-----------------------------GG---AAGATCAG----------------------------------------------------------CAGCAGCAGGCAGAAG---TAGAGGA----------------------------------------------------------------------------------------------------AGTCGAAAACGACTCCGA---------AGATTCATCGG------AATATGAAGAAGTAACAGTCACCGAAAGCAG---CGATGAAGAAGAG |
| dp5 |
3:5960640-5960890 + |
dps_1042 |
ATCAGG------------------------CGTCATCTCCAGTGCGTAC---AGCGACAGCAGCGACGCGAGCCGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTG---GCAGTGACAGCGAGGTAGACGAGGAGAA---T---------------------GAGGAACAGGGGGCA------GAAGA-----------------------------GG---AAGATCAG----------------------------------------------------------CAGCAGCAGGCAGAGG---TAGAGGA----------------------------------------------------------------------------------------------------AGTCGAAAACGACTCCGA---------AGATTCATCGG------AATATGAAGAAGTAACAGTCACCGAAAGCAG---CGATGAAGAAGAG |
| droWil2 |
scf2_1100000004510:2489658-2489926 + |
dwi_441 |
ACCAGG------------------------CAGCATCTCCAGTGCGTAC---AGCCACGACAAGCGCGCGAGCCGAGGTAAGTGTGAATGAGGCAAAT---GAA---------------------------------GAAGAAGATCTGTCCAGTGA---CAGTGGCAGCGAGGAAGAGGACGACGA---A---------------------GAGGAAGATGAT----------------------------------------------------------------------------G---CAGCAGTTGAACAGAA-----------AGAAGTAGTGCCACAGGTACAAG---AGGAGGA----------------------------------------------------------------------------------------------------AAAGGAAAATGAGGATGAAGCTTCAACATCCTCATCAGAGTATGAATACGAAGAGGTTACAGCCACTGAGAGTGA---GTCAGAAGAAGAG |
| droVir3 |
scaffold_12875:19589747-19590036 - |
dvi_7992 |
ATCAGTCGGCGACA------------GCGGCGGCATCTCCAGTGCGTACAACGACAGCGTCGACAGCGCGAGCCGAGGTAAGTGTGAATGAAACAAAT---GAA---------------------------------GAAGAAGGCGTGGCCAGTGACAACAGTGGCAGCAGCGAGACGGAGAGTGA---GACGGGAAGTGAGAGCGAGGCGGAGG---------------------AA-GAAGAGCAGCAGCAGCAGCAGCAGCAG------------------------------GCGC------------AGTTG--------GCGCAGGC---------------ACCCG-------------------------------------------------------------------------------------CTGAGGATCAAGTGGATAACGA------------------------------TTCCAGCTCCGAGTATGAATACGAGGAGGTGACAGCCAGTGAGAGTGA---GC---------AG |
| droMoj3 |
scaffold_6496:6640175-6640509 - |
|
ATCAGGCAGCGACAGCAGCAGCGGCAGCGGCGGCATCTCCAGTGCGTAC---AGCAACGGCGACAGCGCGAGCCGAGGTAAGTGTGAATGAGACAAATGAAGAAGATGAGGAAGCAGATGAGGAAGAAGAAGCAGAAGCAGAAGAGGAAGCCAGTGACAGCAGTGACAGCAGCGAAGCTGAGACTGG---GACCGAAAGTGAGAGTGAGAGTGAGAGTGA---GAC----------------AGAGCAACAGCAGCAGCAGCAGCAG------------------------------GCAG------------AACAA-----------------------------------------------------------------CAC-------CAGCTGCAGCTGCCAGAGC------------------------------------AGGTGGATAATGC------------------------------GACCAGCTCTG------AGTATGAAGAGGTGACAGTCAGTGAGAGTGA---GGAG------AGC |
| droGri2 |
scaffold_15245:17961623-17961945 + |
|
ATCAAG---CGACA------------ACAGCATCATCTTCAGTGCGTACAGCAACAACGGCGACAGCACGAGCCGAGGTAAGTGTGAATGAAACAAATGAAGAAGAAGAAGTGGCAGCAGCAGCAGAAAATGAT---GAAGTAGGTGTGGCCAGTGACGACAGTGGCAGCAGCGACATTGAGAGCGA---A---------------------ACAGAACAA-------------------------------------------------------------------GAGCG------TG---AAC---AAGTTCAAGAGCAAGA--------GTTGCAACCACAAGCAGTAG---CGGTGA---TAAAT----------------------------------------------------------------------GAAGATCAAGTGGACAACGACGACGTCGATGACGC---------------CTCTTCCTCTGAATATGAGTATGAAGAGGTGACAGCGAGTGAGAGCGA---AG---------AG |
| droAna3 |
scaffold_13266:17449042-17449349 - |
dan_2079 |
ATCAGG------------------------CGGCGTCTCCAGTGCGTAACAGTGCAGCAACCGCGACGCGGGCCGAGGTAAGTGTGAATGAGACAAATACAGAA---------------------------------GAAGAAGCCCTGGCCAGCGAAGGCAGTGATAGCGAAGAGGAGGAGGAAGA---A---------------------ACAGAAGAGTCT-------------------------------------------------------------GAAGAAAG------TG---AAG---TAGAACAGGAG--------------------------------GCAG------------GTGTGGCGCCGCAGGAC-------ACGCTTAATGTTCCAGAAG---CTAGCCAGGATGCCTCATCGGGTTCCGAGGTTACAGAAGAGGAGGA------------------------------ATCCTCTTCCG------AATACGAAGAAGAAACAGCAGAAGAAAGCGA---AGAAGAGGAAGAA |
| droBip1 |
scf7180000396735:710631-710938 - |
|
ATCAGG------------------------CGGCGTCTCCAGTGCGTAACAGTGCAGCAACCGCGACGCGGGCCGAGGTAAGTGTGAATGAGACAAATACAGAA---------------------------------GAAGAAGCCATAGCCAGCGAAAGCAGTGACAGCGAAGAGGAGGAGGAAGA---A---------------------ACAGAAGAGTCT-------------------------------------------------------------GAAGAAAG------TG---AAG---TAGAACAGGAG--------------------------------GCAG------------AAGTAAGGCCACAGGAC-------CAGCTTGATGTGCCAGAAG---CTAGCCAGGATGCCTCGTCGGCATCCGAGGTAACAGAGGAGGAAGA------------------------------ATCCTCTTCTG------AATACGAAGAAGAAACAGCAACCGAAAGCGA---AGAAGAAGGGGAG |
| droKik1 |
scf7180000302682:642206-642510 + |
|
CTCAGG------------------------CGGCGTCTCCAGTGCGTAATTCGGCGGCAGCGGCAACGCGAGCTGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTGACAGCAGTGACAGCGAGGAAGAGGAGGAGGA---T---------------------GATGA------GACT------GAGGA-----------------------------GG---AAGAT--------------GA------TG---------------AA--------GTAGAAGA---------------GTCTCCTG------------AGGTC-CTGCCAGAGTTGGCCCAAGGTCTTAGCCCCCCAGAAGTGCCT---GCAGAACCCACATC---CTCTGAACAAACCGAGGAAGAAGA------------------------------GTCTTCCTCGG------AGTATGAAGAAGTAACAGCCACTGAAAGCGA---AG---------AG |
| droFic1 |
scf7180000454044:681139-681515 + |
|
ATCAGG------------------------CGGCGTCTCCAGTGCGTAACAGTGCCGCATCGACGACGCGAGCCGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAATGACAGCAGTGACAGCGAAGAAGAAGTTGAAGA---G---GAAGGGGAGGAGGAGGAGGAGGAGG----------------------AGGAGGAGGAGGAGGAGGAGGAGGAG-G---AAGA--AGAATCAGCAGGGGA------AG---GGG---TAGAACAG--------GTGGAGGA---------------ACCTTCAGTGGGTGTGGTAGAAGAGGCTTCAAGAGAA-------GTTCTTAATCTACCAGAAAAGGCT---TCGAAAGCCACTTC---TTCGAAAGTTCCCGAAGAAGAAGA------------------------------ATACTCTTCCG------AATACGAGGAAGTAACGGCCAACGAAGAGAGTGAAGAAGA---AGAA |
| droEle1 |
scf7180000491228:23783-24078 - |
|
ACCAGG------------------------CGGCGTCTCCAGTGCGTAATAGTGCGGCATCTGCGACGCGGGCTGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTGACTGCAGTGACGGCGAAGAGGAAGAAGAAGA---G---------------------GAAGA--------------------------------------------------------------G-----------GAGGG--------------------------------GGAAGA---------------ATCTGTAGAGGCGGAAATCGAAGAGGCTCCCAGGGAA-------TCTCTTAACCTTCCAGAGGAATCT---CTGCAAGCAACCTC---TTCCGAAGAAACCGAAGAGGAAGA------------------------------ATCCTCATCGG------AATACGAAGAAGAAACAGCCACCGAAAGTGA---AGAG------GAT |
| droRho1 |
scf7180000779618:14865-15163 + |
|
ACCAGG------------------------CAGCGTCTCCAGTGCGTAATAGTGCGGCGACTACGACGCGAGCTGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTGACAGCAGTGACAGCGAAGAAGAGGGGGAGGA---G---------------------GAGGAGGAGGAT----------------------------------------------------------------------------G------------AGGAG--------GGGGAGGA---------------ATCCGTAC------------AGGAAGCTACCGGAAAT-------GTGCTTAACCTTCCAGAAGAAGCT---CTTCAAGCTACATC---TTCCGAAGAAACGGAGGAGGAAGA------------------------------GTCCTCTTCCG------AATACGAAGAAGTCACAGCCACCGAGAGTGA---AGAAGAAGAAGAG |
| droBia1 |
scf7180000302292:1478819-1479105 - |
|
ACCAGG------------------------CGGCGTCTCCAGTGCGTAACAGTGCAGCCTCTACGACGCGAGCCGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTGAAAGCAGTGACAGCGAGGAAGAGGAGGAGGA---G---------------------GAGG----------------------------------------------------------------------------CGGGGGGAG------------AGCAA--------CCGGAAGA---------------ATCTGTTC------------AAGTGGTTCTACAGGAT-------GTGCTTAGTCTA---GGAGAGCCT---CAGCAGCCCACGTC---TTCCGAAGAAACGGAGGAGGAAGA------------------------------GTCCTCCTCCG------AGTACGAGGAAGTAACAGCTAGCGAAACTGA---AG---------AG |
| droTak1 |
scf7180000415991:355332-355651 + |
|
ATCAGG------------------------CGGCGTCTCCAGTGCGTAACAGTGCGGCCTCAACGACGCGAGCCGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTGAGAGCAGTGACAGCGAAGAAGAGGAGGAGGA---G---------------------GAGGATGAGGAGGAA------GAATC-----------------------------GG---TAG---AG-----------GAGGGGGCAG------------AACAA--------GCTGAAGA---------------ATCTGTAG------------AAGGGGCTCCTAGGGAA-------TTGCTTAATCCCCCAGAAGAATCT---CTGCAGCCCACGTC---TTCCGAAGTAACAGAGGAGGAAGA------------------------------ATCGTCCTCCG------AGTATGAAGAAGTAACAGCCACCGAAAGTGA---GGAAGAA---CAG |
| droEug1 |
scf7180000409663:443229-443533 - |
|
ACCAGG------------------------CAGCGTCTCCAGTGCGTAATAGTGCCGCAGCTACGACGCGAGCCGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTGAAAGCAGTGACAGCGAAGAAGAGGAGGAGGA---A---------------------GAAGAAGAGGAT-------------------------------------------------------------------GAGGGTCCAG------------AACAA--------GCAGAAGA---------------ATTTGCAG------------AAGTGGCTTCTAGGGAC-------CGGCTTAACCCACCGGAGGAATCC---CTGCAACCGACCTC---TTCCGAAGTAACGGAGGAGGAGGA------------------------------GTCCTCTTCTG------AGTACGAAGAAGTCACAGCCACCGAAAGTGA---AGAAGAG---CAG |
| dm3 |
chr2R:14885876-14886207 + |
|
ACCAGG------------------------CGGCGTCCCCAGTGCGTAATAGTGCGGCATCCACGACGCGAGCTGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTGAAAGCAGTGACAGCGAAGAAGATGAGGAAGT---T---------------------GAAGATGAAGATGAA---GAAGAAGA-----------------------------GG---TAG---AC-----------GAGGAGGAGG------------AACAA--------GTAGAAGA---------------ATCTGTAGAAGA------AGAAGAGGCTCCCAGGGAA-------TTGCTGAACCTACCAGAGGAAGCT---CTGCAACCGACCTC---ATCTGAAGTTACAGAGGAGGAAGA------------------------------ATCCTCTTCCG------AATACGAAGAAGTAACGGCCACTGAAAGTGA---AGAAGAGCAGGAG |
| droSim2 |
2r:15534852-15535174 + |
dsi_25601 |
ACCAGG------------------------CGGCGTCCCCAGTGCGTAATAGTGCGGCATCCACGACGCGAGCAGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTGAAAGCAGTGACAGCGAAGAAGAGGAGGAGGA---G---------------------GAAGAAGAAGAA---------GATGA-----------------------------GG---TGGAGGAG-----------GAGGAGGAAG------------AACAA--------GTAGAGGA---------------ATCTGTAG------------AAGGGACTCCTAGGGAA-------TTGCTTAACCTACCAGAGGAAGCT---CTGCAACCGACCTC---GTCTGAAGTTACAGAGGAGGAAGA------------------------------ATCCTCTTCCG------AATACGAAGAAGTAACAGCCACTGAAAGTGA---AGAAGAGCAGGAG |
| droSec2 |
scaffold_1:12392163-12392488 + |
|
ACCAGG------------------------CGGCGTCCCCAGTGCGTAATAGTGCGACATCCACGACGCGAGCAGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAAGCGTGGCCAGTGAAAGCAGTGACAGCGAAGAAGAGGAGGAGGA---G---------------------GAAGAAGAAGAA---------GAAGA-----------------------------GGTGGAGGAGGAG-----------GAGGAGGAAG------------AACAA--------GTAGAGGA---------------ATCTGTAG------------AAGGGACTCCTAGGGAA-------TTGCTTAACCTACCAGAGGAAGCT---CTGCAGCCGACCTC---TTCTGAAGTTACAGAGGAGGAAGA------------------------------ATCCTCTTCCG------AATACGAAGAAGTAACAGCCACTGAAAGTGA---AGAAGAGCAGGAG |
| droYak3 |
2R:14307770-14308113 - |
dya_571 |
ACCAGG------------------------CGGCGTCTCCAGTGCGTAGTAGTGCGGCATCCACGACGCGAGCCGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTGAAAGCAGTGACAGCGATGAAGAGGAGGAGGA---T---------------------GTAGAGGAGGAAGATGAAGAAGAAGA-CGAGGAGGAGGAAGATGATGAG----------------GAG-----------GAGGGTGAACAACAAG---TAGAACAA-----------------------------------GTAG------------AAGAAACGGGTAGGGAA-------TTGCTTAACCTCCCGGAAGAGTCA---CTGCAACCGACCTC---TTCTGAAGCAACAGAGGAGGAAGA------------------------------ATCCTCTTCCG------AGTACGAAGAGGTAACAGCCACTGAGAGTGA---AGACGAGCAGGAG |
| droEre2 |
scaffold_4845:9081439-9081776 + |
der_1081 |
ACCAGG------------------------CGGCGTCTCCAGTGCGTAATAGTGCGGCAACTACGACGCGAGCCGAGGTAAGTGTGAATGAGGCAAATACAGAA---------------------------------GAAGAAGGCGTGGCCAGTGAAAGCAGTGACAGCGAGGAAGAGGATGAGGAGGTG------GAGGAGGAGGAGGAAGAAGACG----------------------AAGAGGAAGACGAGGATGAG----------------GAG-----------GAGGGGGAAG------------AACAA--------GCGGAGGA---------------ATCTGTAG------------AAGATACGGGTAGGGAA-------CTGCTTAACCTCCCAGAAGAAGCT---CTGCAGCCGACCTC---TTCTGAAGTCACAGAGGAGGAAGA------------------------------ATCCTCATCGG------AGTACGAAGAGGTGACAGCCACTGAGAGTGA---AGAA------GAG |