
ID:dpe_1078 |
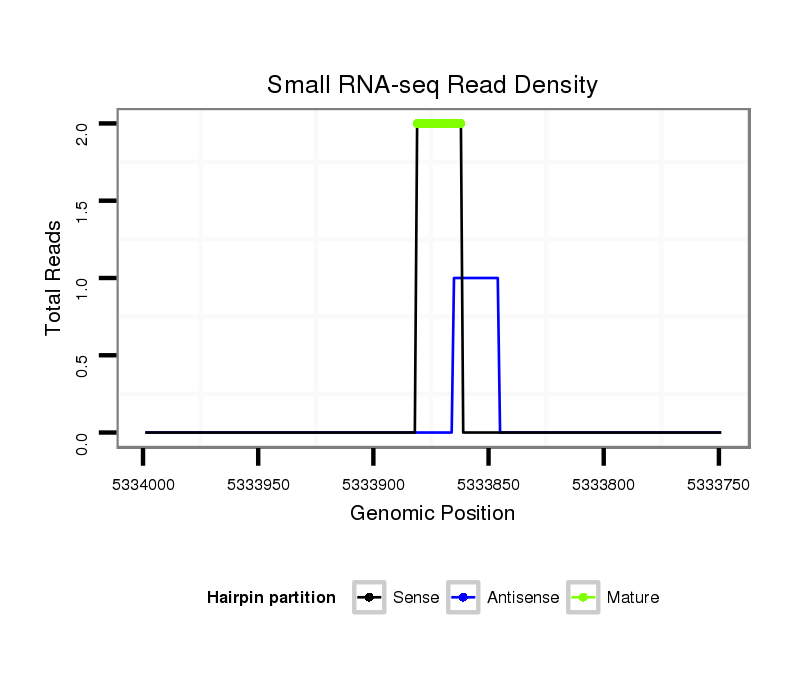
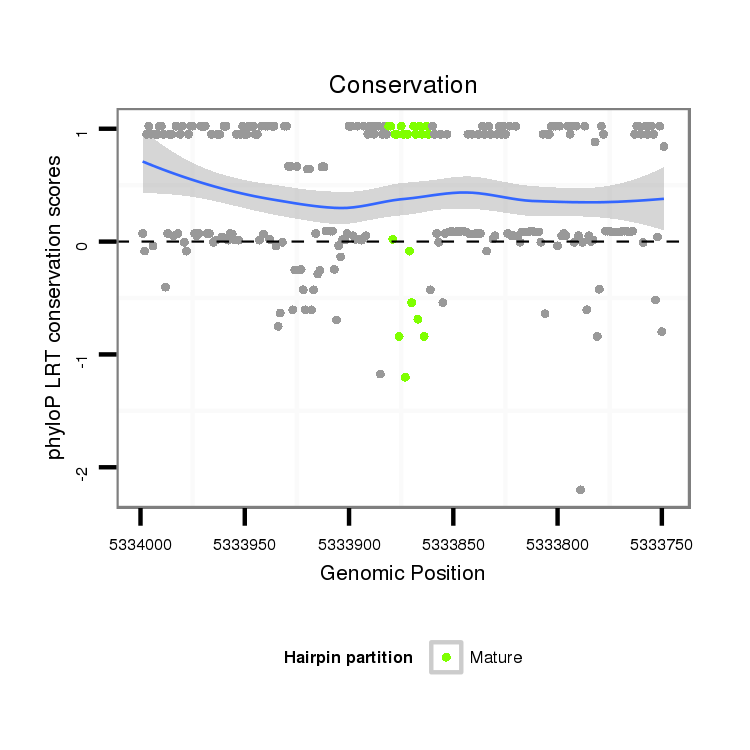
Coordinate:scaffold_2:5333799-5333949 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -24.7 | -24.5 | -24.4 |
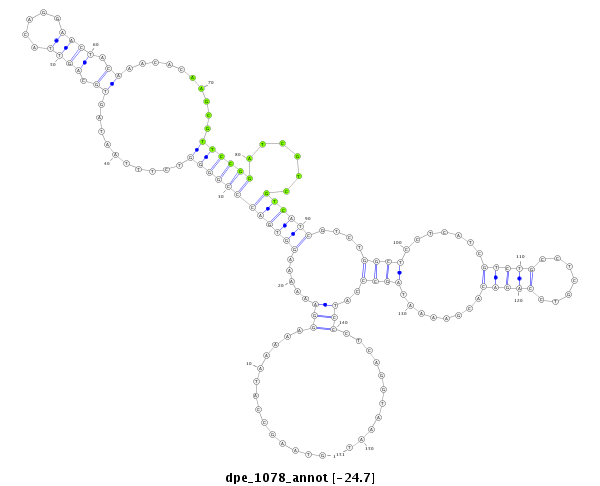 |
 |
 |
exon [dper_GLEANR_10762:1]; CDS [Dper\GL10640-cds]; intron [Dper\GL10640-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CGCTCGCGTTCTCCCAAGGATGGAAGTCATTCGCATCGCAACCATGCCAGGTAAGCCATAAAAAGGAAAAAAGGTGACCCGGGGTCTTTAATAGTGCAGTTACAGGAACTACAAACACAAGCGTTCCGGATCGTCGTCATCGTCTGGCTCCTCATCGTCTGCCTCGTCCAGACACGAAAATAGCCCATCCCTCAGGTAAATACAACAAATGTAGATATCGAGTAACCGGGTATCGACATCAAGTAGGCTAT **************************************************..............(((.....((((((((((((.(((......((.((((.....)))).)).....)))..))))))......))))))....((((.......(((((.......)))))........))))..)))...........************************************************** |
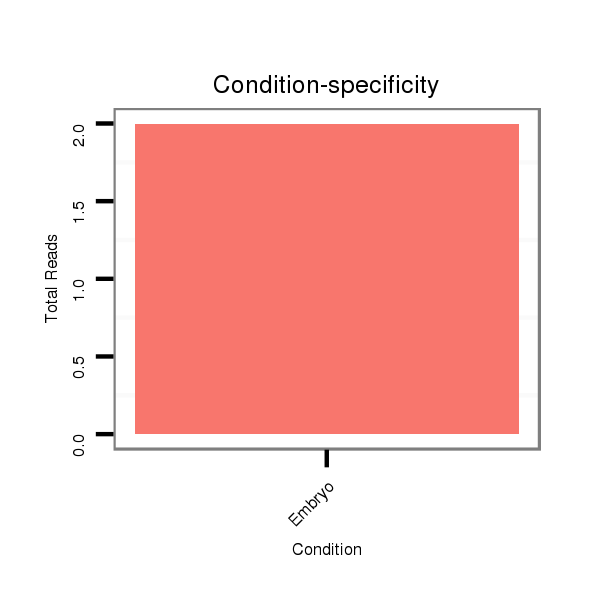
Read size | # Mismatch | Hit Count | Total Norm | Total | M021 embryo |
V111 male body |
V057 head |
V042 embryo |
M042 female body |
|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................AAGCGTTCCGGATCGTCGTC................................................................................................................. | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CCGGATCGTCGTCATCGTCTGGCTCCC................................................................................................... | 27 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................CGGGTATCGACATCAAGTAGGA... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................GGTTGGAAATCATTCGCA........................................................................................................................................................................................................................ | 18 | 2 | 11 | 0.18 | 2 | 0 | 0 | 2 | 0 | 0 |
| ......................................................GCCATAAAAAGGAGAATTGGT................................................................................................................................................................................ | 21 | 3 | 15 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............ATGGTTGGAAATCATTCGC......................................................................................................................................................................................................................... | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ....................TGGAAGTCTTTCGCAGCGC.................................................................................................................................................................................................................... | 19 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................ATGGAAGTCTTTCGCCTAGC.................................................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................ATGGAAGTTATTCGCATAGA.................................................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
|
GCGAGCGCAAGAGGGTTCCTACCTTCAGTAAGCGTAGCGTTGGTACGGTCCATTCGGTATTTTTCCTTTTTTCCACTGGGCCCCAGAAATTATCACGTCAATGTCCTTGATGTTTGTGTTCGCAAGGCCTAGCAGCAGTAGCAGACCGAGGAGTAGCAGACGGAGCAGGTCTGTGCTTTTATCGGGTAGGGAGTCCATTTATGTTGTTTACATCTATAGCTCATTGGCCCATAGCTGTAGTTCATCCGATA
**************************************************..............(((.....((((((((((((.(((......((.((((.....)))).)).....)))..))))))......))))))....((((.......(((((.......)))))........))))..)))...........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V111 male body |
V057 head |
|---|---|---|---|---|---|---|---|
| ......................................................................................................................................GCAGTAGCAGACCGAGGAGT................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| .............................................................................................CGCGTCAATGTGCTTGATC........................................................................................................................................... | 19 | 3 | 5 | 0.20 | 1 | 0 | 1 |
| .......................................................................................................................................................AGATGCAGACGCAGCAGGTC................................................................................ | 20 | 3 | 18 | 0.06 | 1 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droPer2 | scaffold_2:5333749-5333999 - | dpe_1078 | CGCTCGCGTTCTCCCAAGGATGGAAGTCATTCGCATCGCAACCATGCCAGGTAAGCCATA----------------------AAAAGGAAAAA-AGGTGACCCGGG--------GTCTTTAATAGTGCAGTTAC---AGGAACTACAAACACAAGCGTTCCGGATCGTCGTCATCGTCTGGCTCCTCATCGTCTGCCTCGTCCAGACACGAAAATAGCCCATCCCTCAGGTAAATACA-ACAAATGTAGATAT----C------------------------------GAGTAACCGGGTATCGACATCAAGTAGGCTA-------------------T |
| dp5 | 3:5132554-5132804 - | CGCTCGCGTTCTCCCAAGGATGGAAGTCATTCGCATCGCAACCATGCCAGGTAAGCCATA----------------------AAAAGGAAAAA-AGGTGACCCGGG--------GTCTTTAATAGTGCAGTTAC---AGGAACTACAAACACAAGCGTTCCGGATCGTCGTCATCGTCTGGCTCCTCATCGTCTGCCTCGTCCAGACACGAAAATAGCCCATCCCTCAGGTAATTACA-ACAAATTTAGATAT----C------------------------------GAGTAACCGGGTATCGACATCGAGTAGGCTA-------------------T | |
| droFic1 | scf7180000453778:756006-756259 + | TCCTCCCGTTCCTCCGAAGAGCGAAACTATTTACATCGCAAATTCGACAGGTAAGCAAAA----------------------ATAACTTAAAATACATAGAGCGCTGTGTTGAGA--------GTCTCGATTGCAGTAGTAGCTACAAGCACAAGCGGTCTGCTTCCTCCTCGTCAGCAAG---------GACAACTTCCTCTGGACACAAGGACC---------GCCGGTAAATGTGTACACACATATGTG----AGCCCCTCCTAAACGATACCTATTTTGACAGCAAG--------------CATCGAGTAGGCTT-------------------T | |
| droRho1 | scf7180000779318:325114-325381 - | TCCTCGCGTTCCTCCGAAGAGCGAAACTATTTACCACGCAAATTTGACAGGTAAGCAAAA---------CATTATTTACATTAAAAGACCAA--------------GTGTTGAGAATTT--------TAATTGTAGCAATAGCTACAACCACAAACGGTCGGCGTCATCCTCATCAGCCAG---------GACAACTTCCTCCGGACACAAGGACC---------GCCGGTAAATGTGTACACACAGATGCATGTGTG-CCCCCCTACACGATAATT--TTTGACACCGAG--------------CATCGAGTAGTCTG-----GCAGTTTAGAAAAGT | |
| droEug1 | scf7180000409462:2345499-2345765 + | TCCTCGCGTTCTTCCGAAGAGCGAAACTATTTACATCGTAAATTTGACAGGTAAGCAAAATGCAATATAAAATATGTACCTAAAAAGTTAAA--------------GTGTTGAGA---------ACTCAATTGCAGCAGTAGCTACAAACACAAGCGCTCCGCGTCATCATCGTCAGCCAG---------GACAACTTCCTCCGGACACAAGGACC---------GCAGGTAAATGTGTACACACTTAAGTAT----TAAACTCCTAAACGATACCTA-TTTGACATCAAG--------------CATCGAGTAGTTTTGGCTGACA------------ |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
Generated: 06/08/2015 at 03:42 PM