
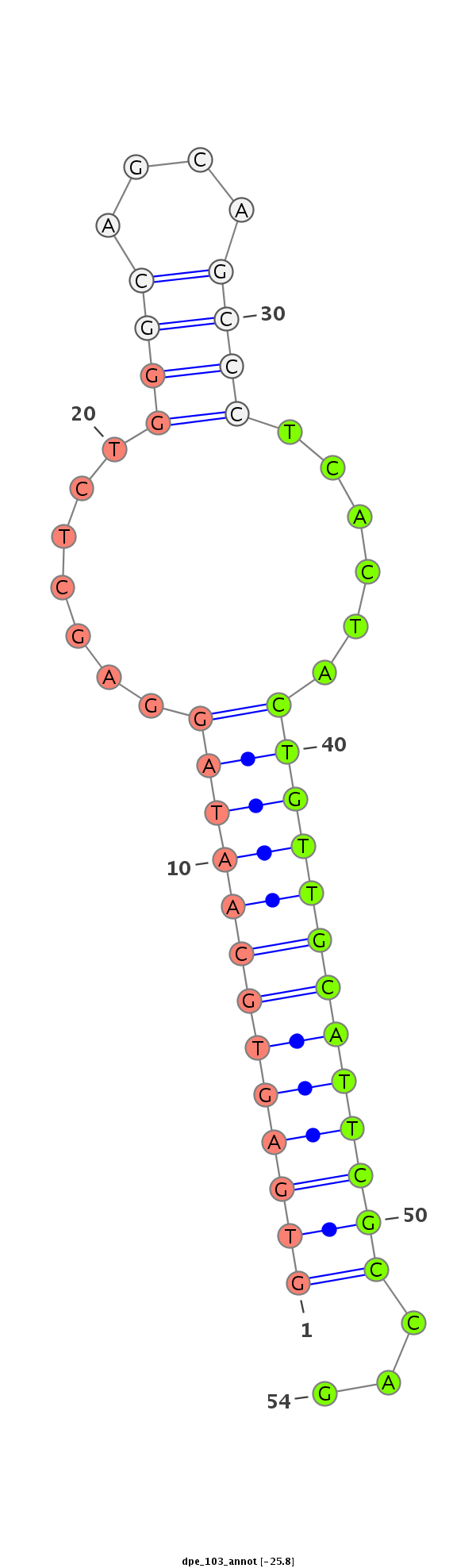
ID:dpe_103 |
Coordinate:scaffold_31:765354-765407 - |
Confidence:confident |
Class:Mirtron |
Genomic Locale:intron |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
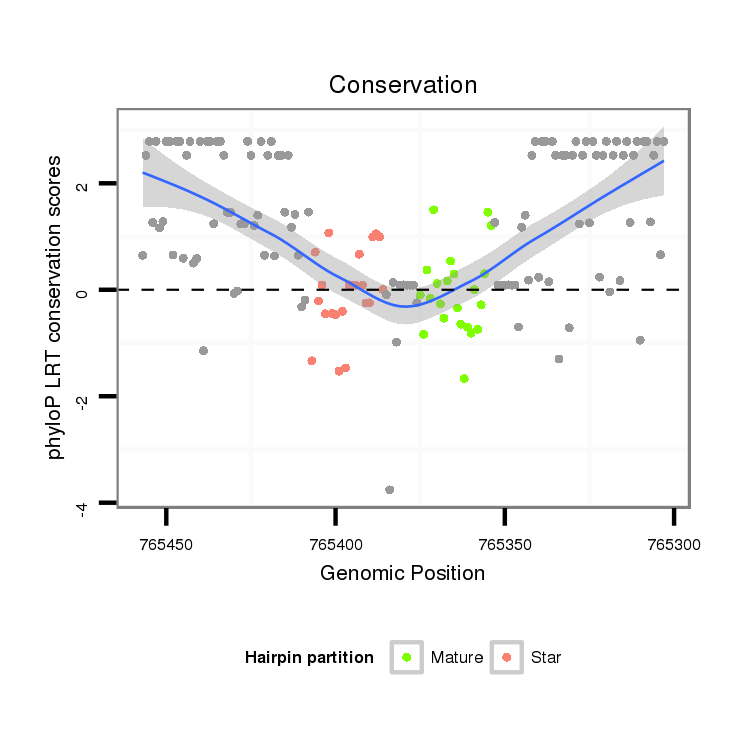
| -25.8 |
 |
intron [Dper\GL10088-in]; CDS [Dper\GL10088-cds]; CDS [Dper\GL10088-cds]
No Repeatable elements found
|
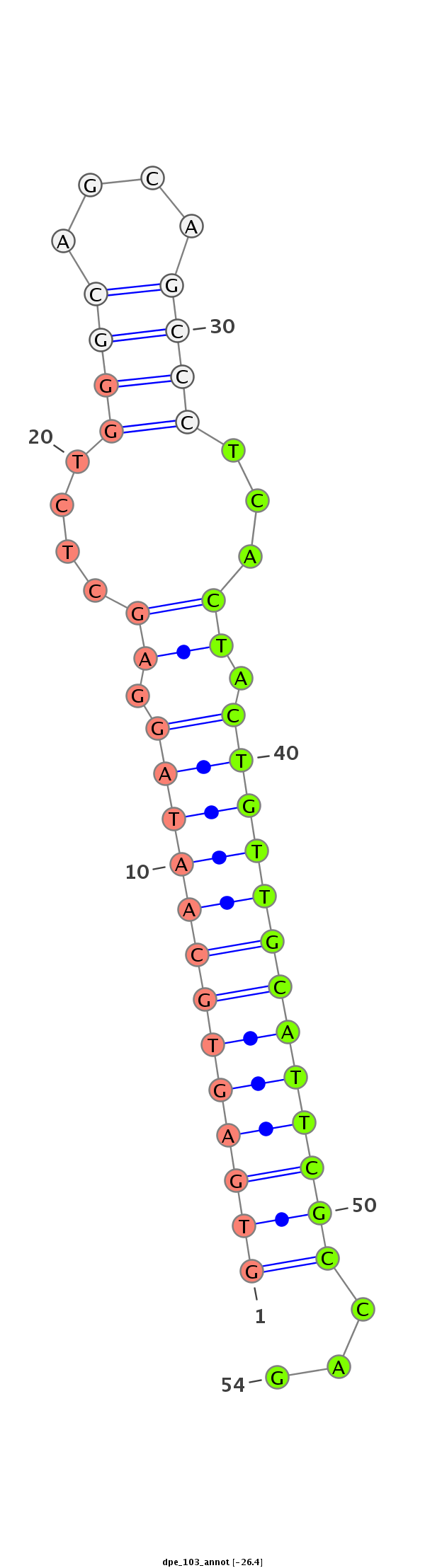
AGAGTCCAAGAACCATCTGAACTACTACACCAGCAAGCAGCCAGCTGCAGGTGAGTGCAATAGGAGCTCTGGGCAGCAGCCCTCACTACTGTTGCATTCGCCAGCACGACAAGACGACAAGTCGGCGCAGGAGAGCGACGACGAGGTGAAGGAC
**************************************************(((((((((((((.((....((((....))))...)).)))))))))))))...************************************************** |
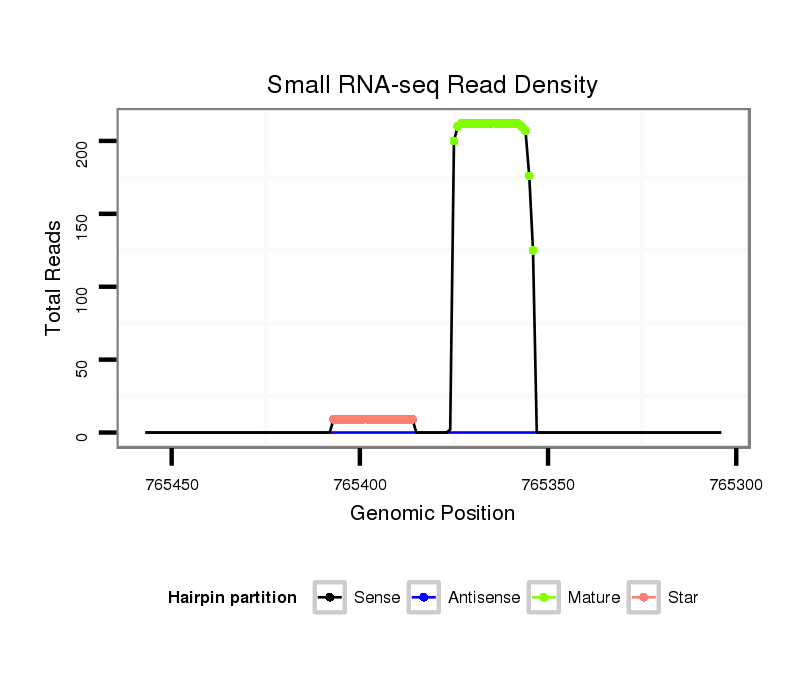
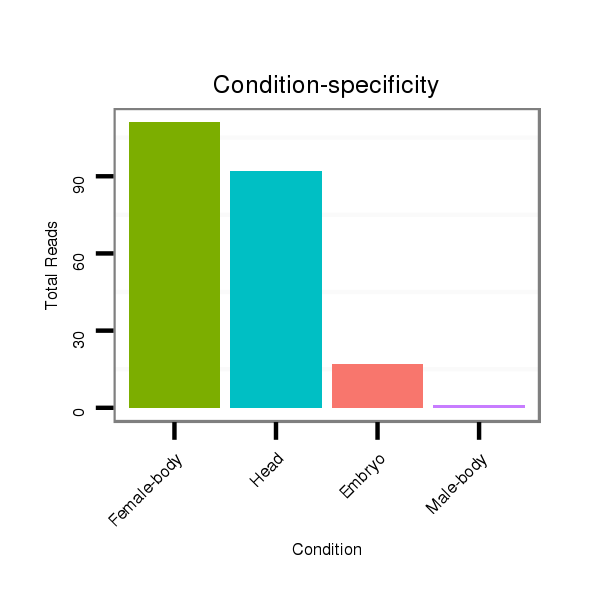
Read size | # Mismatch | Hit Count | Total Norm | Total | M042 female body |
V057 head |
V050 head |
M021 embryo |
V111 male body |
|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................TCACTACTGTTGCATTCGCCAG.................................................. | 22 | 0 | 1 | 119.00 | 119 | 69 | 17 | 22 | 11 | 0 |
| ..................................................................................TCACTACTGTTGCATTCGCCA................................................... | 21 | 0 | 1 | 45.00 | 45 | 26 | 12 | 5 | 2 | 0 |
| ..................................................................................TCACTACTGTTGCATTCGCC.................................................... | 20 | 0 | 1 | 29.00 | 29 | 5 | 16 | 4 | 4 | 0 |
| ..................................................GTGAGTGCAATAGGAGCTCTGG.................................................................................. | 22 | 0 | 1 | 9.00 | 9 | 0 | 5 | 3 | 0 | 1 |
| ...................................................................................CACTACTGTTGCATTCGCCAG.................................................. | 21 | 0 | 1 | 5.00 | 5 | 4 | 0 | 1 | 0 | 0 |
| ...................................................................................CACTACTGTTGCATTCGCCA................................................... | 20 | 0 | 1 | 5.00 | 5 | 5 | 0 | 0 | 0 | 0 |
| ..................................................................................TCACTACTGTTGCATTCGCCAGT................................................. | 23 | 1 | 1 | 3.00 | 3 | 0 | 1 | 2 | 0 | 0 |
| ..................................................................................TCACTACTGTTGCATTCGC..................................................... | 19 | 0 | 1 | 3.00 | 3 | 1 | 0 | 2 | 0 | 0 |
| ..................................................................................TCACTACTGTTGCATTCGCCT................................................... | 21 | 1 | 1 | 2.00 | 2 | 0 | 1 | 1 | 0 | 0 |
| ..................................................................................TCACTACTGTTGCATTCG...................................................... | 18 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 |
| .................................................................................CTCACTACTGTTGCATTCGCCA................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................TCACTACTGTTGCCTTCGCCA................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................ACTACCGTTGCATTCGCCAG.................................................. | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................TCACTACGGGTGCATTCGCCA................................................... | 21 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................GTGAGTGCAATAGCAGCCCTGC.................................................................................. | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................TCACGACTGTTGCATTCGCCAT.................................................. | 22 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................TAAATACTGTTGCATTCGCCAG.................................................. | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................ACTACTGTTGCATTCGCC.................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................TCACTACTGTTGCATTCCCCAG.................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................TCACTACTGTTGCATTCGCCCT.................................................. | 22 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................TCACTACTGTTACATTCGCA.................................................... | 20 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................TCACTACTGTTGCATTCGCCAT.................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................TCACTACTGTTGCATTCGCCG................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................CTCACTACTGTTGCATTCGCC.................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................CACTACTGTGGCATTCGCCAG.................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................TCACTACTGTTCCATTCGCCAG.................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................TCACTACTATTGCATTCGCCA................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................TCACTACTGTTGCATTCGAC.................................................... | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................TCACTAATGTTGCATTCGCCA................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................TTACTGTTGCATTCGCCAG.................................................. | 19 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................TCACTACTGTTGCATTCGCCAGAC................................................ | 24 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................AGTCGGCGCCGGACAGCGACTAC............ | 23 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................TCACTACTGTTGCAGTCGCT.................................................... | 20 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................TCACTACCGTTGCATTCGCCA................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................TCACTACTGATGCATTCGCCA................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................TCACTACTGTTGCATTCGCCAC.................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................ACTACTGTTGCATTCGCCAG.................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................TCACTACTGATCCATTCGCCAA.................................................. | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................TCACTACTGTTGCATTCCCCA................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................TCACTCTGGTTGCATTCGCCAG.................................................. | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................TCACTACTGTTGCATTCGCCAGA................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................TCACTACTGTAGTATTAGCCA................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................CGACGACGATGTGAAGGA. | 18 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................TCACGACTGTGGCATTGGCCA................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
|
TCTCAGGTTCTTGGTAGACTTGATGATGTGGTCGTTCGTCGGTCGACGTCCACTCACGTTATCCTCGAGACCCGTCGTCGGGAGTGATGACAACGTAAGCGGTCGTGCTGTTCTGCTGTTCAGCCGCGTCCTCTCGCTGCTGCTCCACTTCCTG
**************************************************(((((((((((((.((....((((....))))...)).)))))))))))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V057 head |
V111 male body |
|---|---|---|---|---|---|---|---|
| .....................................................................ACCCGTCATCGTGAGTAATG................................................................. | 20 | 3 | 5 | 0.20 | 1 | 1 | 0 |
| ...........TGGTAGACTCGATGAGGTCG........................................................................................................................... | 20 | 3 | 15 | 0.13 | 2 | 1 | 1 |