| ACGT----------TTG----AA--ACTG------CA----------------------------------------T-GG-----AGAATGCGGAAACAAAC---ACAA----------ACCATACACA------------------------CGTACTT---------------------------ATTACAC---------------A-------CA--C--ACATACATAC-----------------A----------------CA------CACACACACACACACACACACA---------------------------------C------------AC-------ACACAC-----AC------------A----C-----------------------ACAC------A--------CACACACACACACACACACACA--------------CACACACAAA-------------------CA-----CACAGACATGGACGGAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAACAC | Size | Hit Count | Total Norm | Total | M020
Head | M045
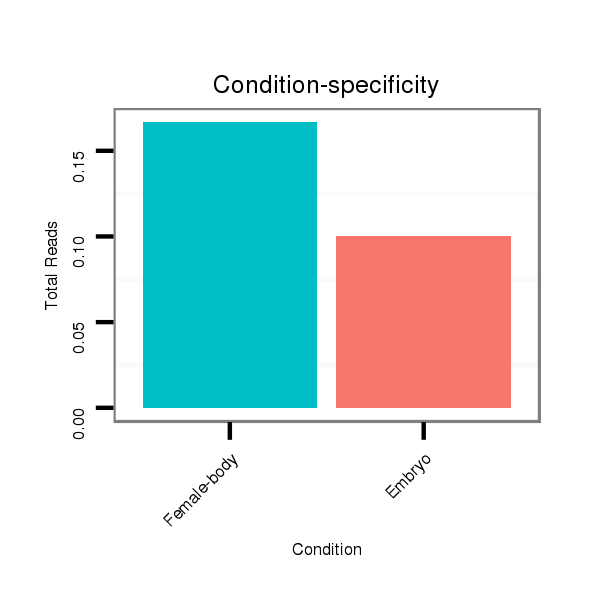
Female-body | V117
Male-body | V118
Embryo | V119
Head |
| ................G----AA--ACTG------CA----------------------------------------T-GG-----AGAATGC................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 19 | 1 | 6.00 | 6 | 0 | 1 | 2 | 3 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................A-----CACAGACATGGACGGAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------...... | 18 | 1 | 4.00 | 4 | 3 | 0 | 0 | 1 | 0 |
| .....................AA--ACTG------CA----------------------------------------T-GG-----AGAATGC................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 18 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 | 0 |
| ...............TG----AA--ACTG------CA----------------------------------------T-GG-----AGAATGC................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 | 0 |
| ACGT----------TTG----AA--ACTG------CA----------------------------------------T-GG-----A....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 | 0 |
| ..............TTG----AA--ACTG------CA----------------------------------------T-GG-----AGAATGC................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 21 | 1 | 2.00 | 2 | 0 | 0 | 1 | 1 | 0 |
| ....................................A----------------------------------------T-GG-----AGAATGCGGAAACA.......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 18 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AA-------------------CA-----CACAGACATGGACGGAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A..... | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CACACAAA-------------------CA-----CACAGACATGGACG............................................................................................................................................................................................. | 24 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AA-------------------CA-----CACAGACATGGACGGAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAC.. | 25 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................AAC---ACAA----------ACCATACACA------------------------CGTA................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 21 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................T-GG-----AGAATGCGGAAACAAA........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 19 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................AAAC---ACAA----------ACCATACACA------------------------CGT................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..GT----------TTG----AA--ACTG------CA----------------------------------------T-GG-----AGAA.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AAA-------------------CA-----CACAGACATGGACGGAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------...... | 22 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..GT----------TTG----AA--ACTG------CA----------------------------------------T-GG-----AGA..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..............TTG----AA--ACTG------CA----------------------------------------T-GG-----AGAA.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................CA----------------------------------------T-GG-----AGAATGCGGAAACA.......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................A-------------------CA-----CACAGACATGGACGGAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A..... | 21 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CA-----CACAGACATGGACGGAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAACAC | 25 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................CATACACA------------------------CGTACTT---------------------------ATTACAC---------------............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 22 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................A-----CACAGACATGGACGGAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A..... | 19 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..GT----------TTG----AA--ACTG------CA----------------------------------------T-GG-----AGAATGCGG............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................T-GG-----AGAATGCGGAAACAAAC---ACAA----------ACC................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ....................................A----------------------------------------T-GG-----AGAATGCGGAAACAAA........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 20 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CACAGACATGGACGGAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAA... | 20 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................CATACACA------------------------CGTACTT---------------------------ATTACA............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 21 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AA-------------------CA-----CACAGACATGGACGGA........................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..GT----------TTG----AA--ACTG------CA----------------------------------------T-GG-----AGAATGC................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................A----------------------------------------T-GG-----AGAATGCGGAAACAAAC---ACAA----------ACC................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .CGT----------TTG----AA--ACTG------CA----------------------------------------T-GG-----AGA..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AA-------------------CA-----CACAGACATGGACG............................................................................................................................................................................................. | 18 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CA-----CACAGACATGGACGGAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A..... | 20 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GACATGGACGGAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAACAC | 19 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................AAC---ACAA----------ACCATACACA------------------------CGT................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................................................................CACACACAAA-------------------CA-----CACAGACATGGACG............................................................................................................................................................................................. | 26 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................ATGCGGAAACAAAC---ACAA----------ACCATA................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 24 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................CA----------------------------------------T-GG-----AGAATGCGGAAACAAAC---A................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............TG----AA--ACTG------CA----------------------------------------T-GG-----AGAATGCGGAAACA.......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................T-GG-----AGAATGCGGAAACAAAC---AC.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAAA-------------------CA-----CACAGACATGGAC.............................................................................................................................................................................................. | 19 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................A-----CACAGACATGGACGGAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAC.. | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................TACACA------------------------CGTACTT---------------------------ATTACAC---------------A-------CA--C--ACA.......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................ACATACATAC-----------------A----------------CA------CACAC.................................................................................................................................................................................................................................................................................................................................................................................................................................... | 18 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |