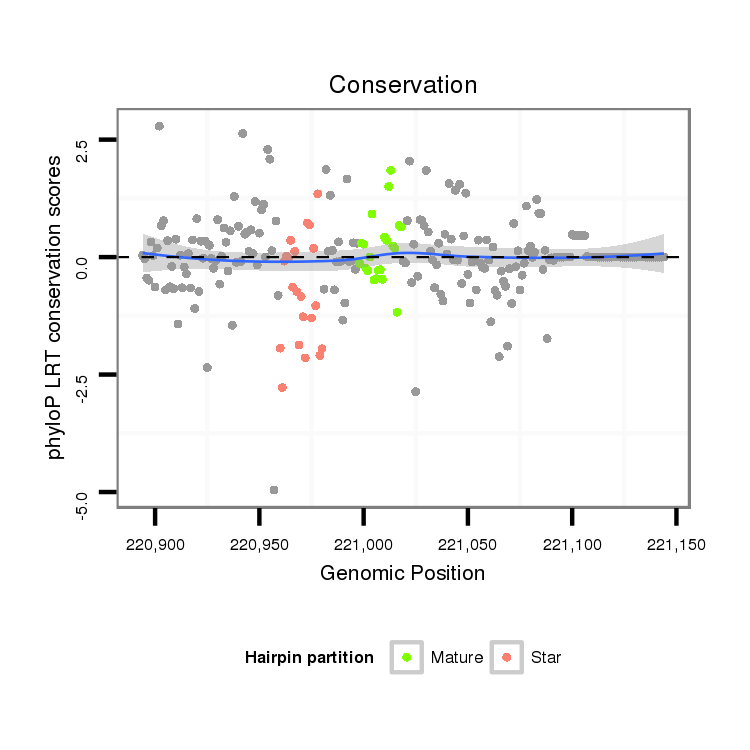
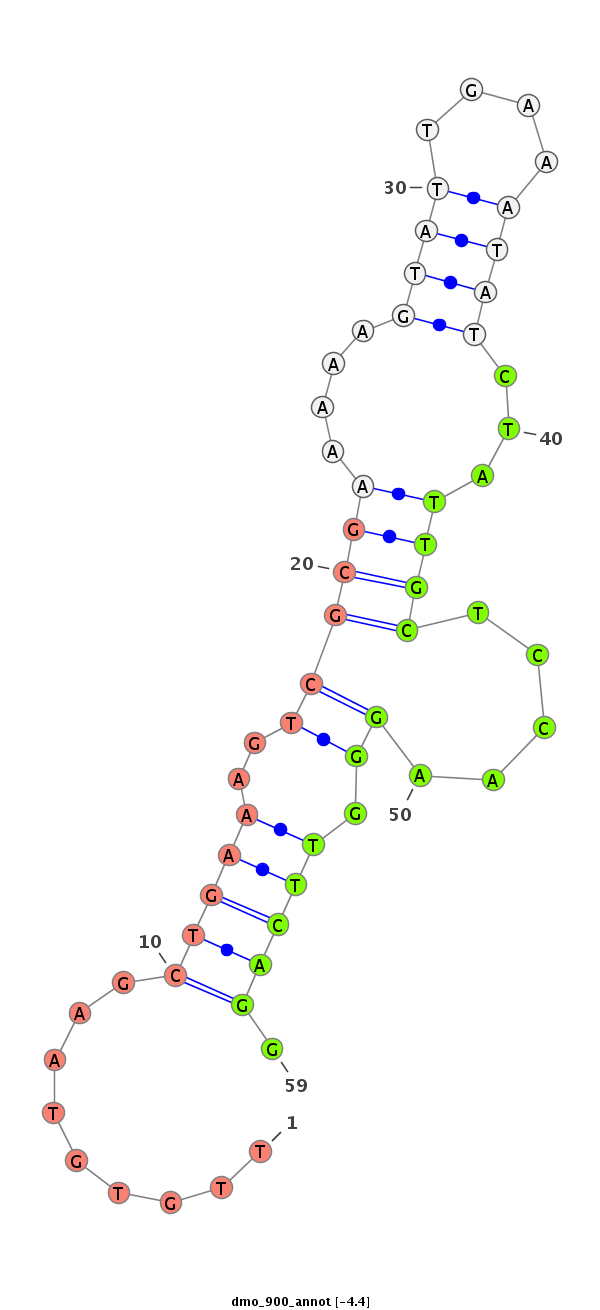
| droMoj3 |
scaffold_6496:220894-221144 + |
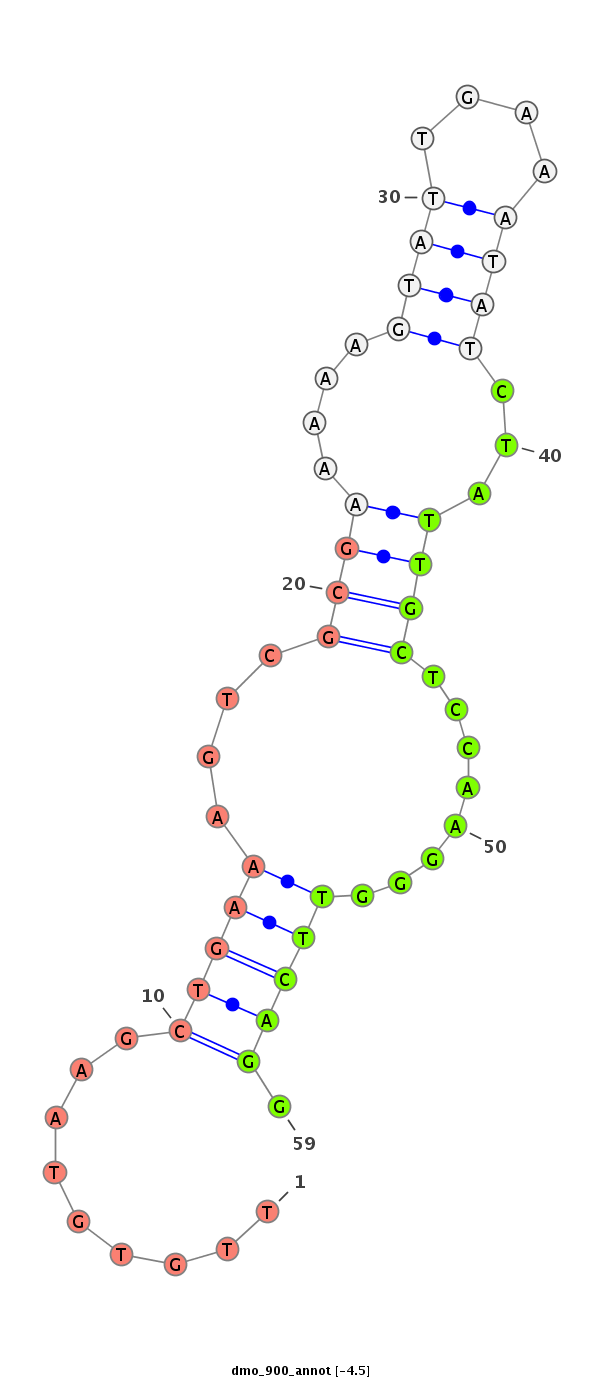
dmo_900 |
TGTATGCGTGTGTGTGTATGTATGC----------------CTGTGTTTGCGTACGTGCGTGTGTGTGTGTGTGCGTGTT-----------------------TCT-------------T-----------G--T--GTAAGCTGA-----------AAGT-----------C-----------------------------------------GCGAAAAAGTATTG-------------------------------------------------AAATATCTATTGCT----------CCAAGGGTTCAGGGGATCTCGTAGTCGA---GCAGTCTCGACTAGAGC-----------------------------------------------------------------------------------------------TTTCTTACTTGTTATATT--------ATCTCTT--A-----------TTTGTCTCGCCCATGCAGGTTGTTGGCGCCAC-------------------------------------GGAGGAGGTGAACAGCGCCACTGAGATGCACGAGGG |
| droVir3 |
scaffold_12970:3028496-3028723 - |
|
AGCGCG-GTGTTTCTATGCGTTTAT------------GC--GTGCGTGCGTGTGTGTGCATGTGTGAGTATGTGTGTGTA-----------------------TCT-------------G---------TAG--AAAG--AAG-----------------------------------------CAAGAATATT-----------TTTAAC------------------------GGCAGCTCGG--------------------------------------------CG----------CTGTGGGTTCAGGGTATCTTATAGTCGG---GCACTCACGGCCAGAGT-ACTATTAGTTGTATTTTGTTGGTTT----TTAAGTTTTTTGAC---------------------------------------------------TGCT--TTGGCTGAGGCA--------TGCTTAA--G-----------CTGG-------------------TGGCGCCAC------------------------------------------------------------------------- |
| droGri2 |
scaffold_14906:13053836-13054029 - |
|
TGTATGTATGTATGAATATTCACGT----------------GTTTGTCGGC-TGCGC---------------------TC-----------------------ACC-------------TT--TTG-----G--TGC---AAAACTTTTTAGAGGGAAATT-----------TTCCAACTTGAACTAGAATATT---------TGTTTAAC------------------------------------------------------------------------------------------AGCAGGGTTCAGGGTATATCGTAGTCGA---CCTCACTCGACTACAGTTACTGTTACTTGTTT-ATGTTCGTCTGTCGTGTA----------G-------------------------------------------------------------------------------------------CTTG-----------TCAATG------------------------------------------------------------------------------------------ |
| droWil2 |
scf2_1100000004401:654384-654488 + |
|
TGTATGTGTGTGTGTGTGTGTGTGT----------------GTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTG-------------------------------------------------TGG--TGTG--AGATAT-----------ATTT-----------C-----------------------------------------GA-CAAACATATGA-------------------------------------------------ATGTATCTA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T------------------------------------------------------------------------------------------------------- |
| dp5 |
XR_group6:12953152-12953264 - |
|
TCTGTGTGTGTGTGTGTGTGTGTGT----------------GTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGCT-----------------------TTG-------------G-----------A--T--GTGTG--------------------------------------------------------------CCTGCAAGCAGTGCATACATATGT-------------------------------------------------ATGTGTTTACA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTT------------------------------------------------------------------------------------------------------- |
| droPer2 |
scaffold_14:343983-344119 + |
|
TGTGTGTGTGTGTATGTATGCGTTT----------------GTGGGAGGGGTTATGTTTG---------------------------------------------GGGTGTATATGGGGTA--TATGTATGA--TTAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGAGGGCTCTCTTTCTAGGCTCAGTCGCAGTCTACAAATCTTAACTAACGTTTTTT-----------------------GTTT-------------TGTT------------------------------------------------------------------------------------------------------- |
| droAna3 |
scaffold_13335:1859816-1859997 - |
|
GGTGTGTGTGCTCAGGTAGGGGTGT----------------ATGTGTGTGCGTAGGGGCGCGTGTTTGCGTAGGTGTGTGCT---------------------TTT-------------TGTATCT-----G--TGTG-------------------AAGTGTATCGAGTGTA------------------------------------------TTGAA--------CTCTTGCTGCATTTTTGCACAATTTCGACTTG---------------------------------------------------------TCTT-----------------------------------TTCTTTTT-ATATTCATTTC----------------------------------------------------------------------------TT--------TTAATTTTATTTTTT--A-----------TTTATTT-------------------------------------------------------------------------------------------------- |
| droBip1 |
scf7180000396434:275106-275205 + |
|
TGTGAGAGTGTGTGCATGTGTGTGT----------------TTGTATATATATGTGTGTGTCAGTGTGTTTGTGTTTGTA-------------------------------------------------TGG--T----GGAATGG------------------------------------------------------------------------------------------------------------------------------------------------------------------TAATGGAAATAAGATAGTAAAAA----A------------------------------------------------------------------------------------------------------------------------------------------------------------T------------------------------------------------------------------------------------------------------- |
| droKik1 |
scf7180000302398:414012-414128 + |
|
TGTGTGTGTGTGTGTGTGTGTGTGT----------------GTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGT-----------------------------TG-------------TG--TATGTGTACGCATTA--A--------------------------------------------------------------------------------------------------------------------------------------------------------------------AGGTAAAATATGTTTTGTTATCAATG----TTTTC-------------------------------------------------------------------------------------------------------------------------------------AAT-------------GCTT------------------------------------------------------------------------------------------------------- |
| droFic1 |
scf7180000454077:228513-228630 - |
|
TGTGTGTGTGTGTGTGTGTTTCTCT----------------CTCTCTTTCTGTGCGTGTATGTGTGGGTGTGTGTGTGGT-----------------------TCA-------------G-----------G--T--GG-------------------------------------------------AAAAGC---------GGCTAAAAAGCGGGGAAACGCGAAG-------------------------------------------------AAAAGCCCG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCACTC------------------------------------------------------------------------------------------ |
| droEle1 |
scf7180000490982:807510-807689 + |
|
TCAATGAGTGTGTGTGTGTGTGTGT----------------GTGTGTGTGTGTGTGTGTGTGTTTGTATT---------------------------------TCA-------------G-----------T--T--ATAAATTGG-----------AACC-----------A-----------G--------------------CCGAAAAAAGTTGAA--------CAGATGTCGTTTTCTTTACCAGTTATCGTT--------------GGAAGA------------------------------------------------------GAAATCTCATCTAAG-T-----------------------------------------------------------------------------------------------TGCTGTATTTACAAGGTG--------TGTTTTT--G-----------TTTGCCTCGCCC--------------------------------------------------------------------------------------------- |
| droRho1 |
scf7180000778552:5178-5318 + |
|
G-CGTATGTGTGTATGTATGTAAGC----------------ATATACATATGTATGTATTTACAT---------------------------------------T--------------------------G--T----A--------------------------------------------CAC-AGTTGC---------AATTGTTT------------------------------------------------------------------------------------------AGCTGAGTAACGGGTATCTAATAGTCGAGG----CACTCGACTATAGC-----------------------------------------------------------------------------------------------GTTCTTTCTTGTTTTGTT--------TTATTT-------------TCTT------------------------------------------------------------------------------------------------------- |
| droBia1 |
scf7180000299898:36646-36766 - |
|
TGTGTGTGTGTGTGTGTGTGTGTGT----------------GTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTG-------------------------------------------------TGG--TGGG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------G-------------------------------------------------------------------------------------------GGGG-----------GGGAGG---------------GTTCGAACCCGTCTGAAAAATACGTTTCCTCGAGGTCTC------------------------------------ |
| droTak1 |
scf7180000412666:10972-11162 - |
|
TGTCTGTCTGTCTGTCTGTCTGTCT----------------GTCTGTCTGTCTGTCTGTCTGTCTGTCTGTCTGTCTGTCTG-----------------------------------------------TCG--A----AATTAAA-----------AAGT-----------T----ATACGCAA---------TCAAAAAATTGTATATA----------------------T-CCATCTCCCT--------------------------------------------CGCACTGCCTTTAGCAGAGTAACGGGTATTAGATAGTCGGGGCACAAACCCGACTATAGC-----------------------------------------------------------------------------------------------GTTTTCTCTTG------------------------------------TT------------------------------------------------------------------------------------------------------- |
| droEug1 |
scf7180000409040:52234-52373 - |
|
TGTGTGTGTGTGTGGGAAAATGCGTCTAAGTGTGTTTGTTTGCATGCATGTGTGTGTATATCTGTGTGTGTGTGTGTGCGTGTATGGCCAGAGCGCCAATATGTAT-------------G-----------C--T--ATATATAGA-----------GAAA-----------A-----------C----------------------------------------------------------------------------------------------------------------------------TTAGGAGATCTTGAAG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| dm3 |
chrX:8638799-8638909 - |
|
TGCGTACGTGTGTGCGCGAGTGTAT----------------CTGTGTGTGCGAATGTATCTGTGTGTGCGAGTGTGTGTC-----------------------TCA-------------TG--TTT-----G--CGC---AAATCGTTAGAGCG-------------------------------------------------------------AAGAG--TTGTGT-------------------------------------------------AAATATTTG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T------------------------------------------------------------------------------------------------------- |
| droSim2 |
2r:8841766-8841887 - |
|
CAGGAAAGTAATGA-AATTTCGCGC----------------TCGCA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTCTCACTAGCTGAGTAAGGGGTATCTGATAGTCGGCG----AACTCGACAATAGC-----------------------------------------------------------------------------------------------TTTCTCTCTTGTTTTAAT--------ATCTCTTTGATTTTTGTTATCCA------------------------------------------------------------------------------------------------------- |
| droSec2 |
scaffold_10:2912021-2912124 + |
|
TGTGTGTGTGTGTGTGTGTGTGTGT----------------GTGTGTGTGTGTGTGTGTGTGTGTGTGTGTATGTGTGTGTGTG---------------------------------------------TGG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTGA--------------------------------------------------------------------------------------AC--------------AGGCGCAGTCGCTGT--GAAAGT---AGCAA----------------------------------------------------A------------------------------------------------------------------------------------------------------- |
| droYak3 |
v2_chr3L_random_067:5434-5570 + |
|
TACATACATATGCATATATACATAT----------------GTATGTATATGAATGCATATTTATT-----------------------------------------------------------------G--T----T--------------------------------------------------TTTT---------AAATAATC------------------------------------------------------------------------------------------TGCTGAGTAACGGGTATCTAATAGTCGGGG----AACTCGACTATAGC-----------------------------------------------------------------------------------------------GTTCTCTCTTATTAAATT--------TTCTCT-------------TATT------------------------------------------------------------------------------------------------------- |
| droEre2 |
scaffold_4770:9318743-9318933 + |
|
TGTATGTATGTATGTATGTATGTAT----------------GTATGTATGTATGTATGTATGTATGTATGTACATATGTATGTTTGG-----------------TC-------------A---------TA------ATCAGCAGC-----------AAGT-----------T----GCTACCGA---------AAA------------------CCAAA---CGAT-CTTTGGTCACCTTC---TGGAATTCCTGTTCGCCTGGTAATTTAAACCGA------------------------------------------------------AAAATC------------------------------------------------------------------------------------------------------------------TC--------ACAATTTTGTCTCCA--A-----------ATTGTCTCGC----------------------------------------------------------------------------------------------- |