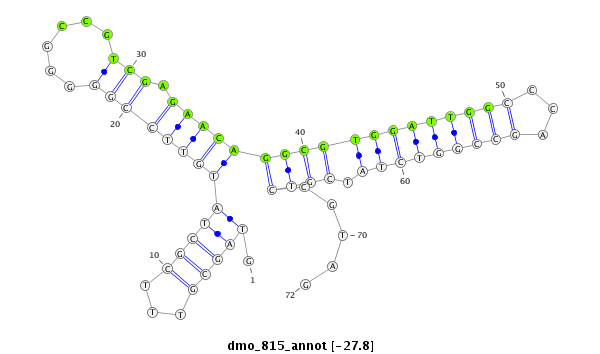
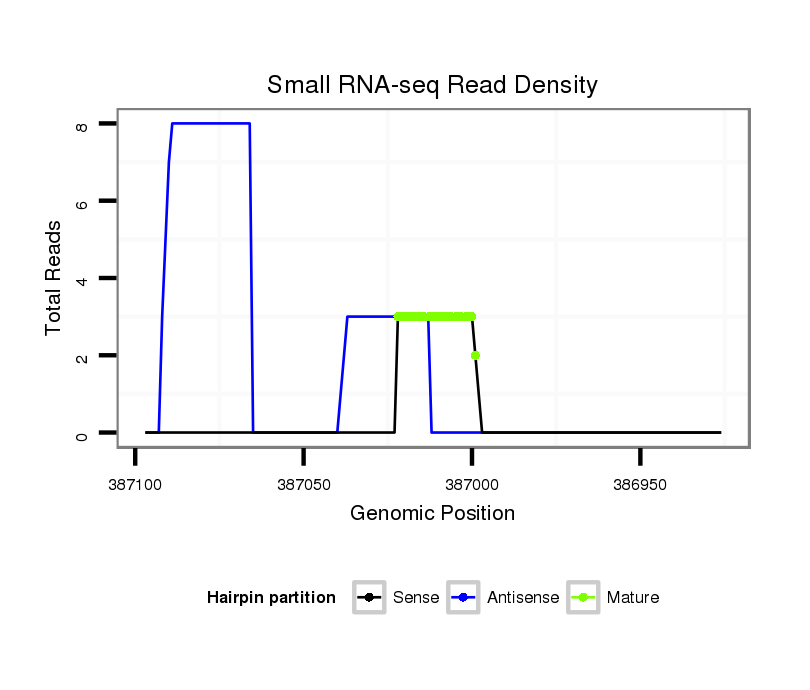
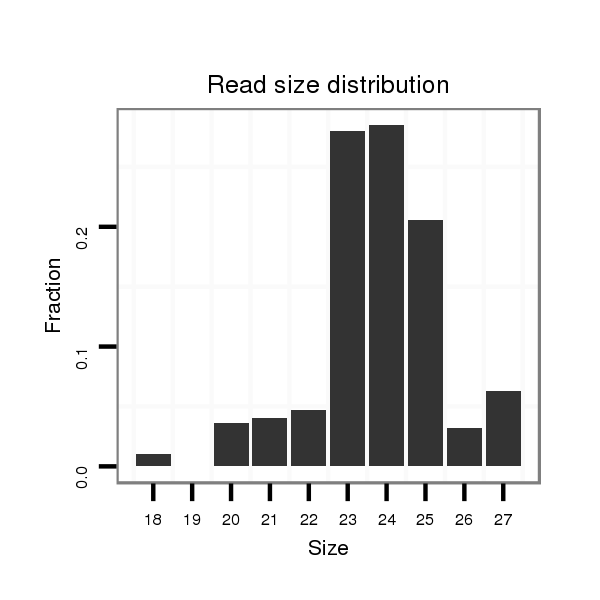
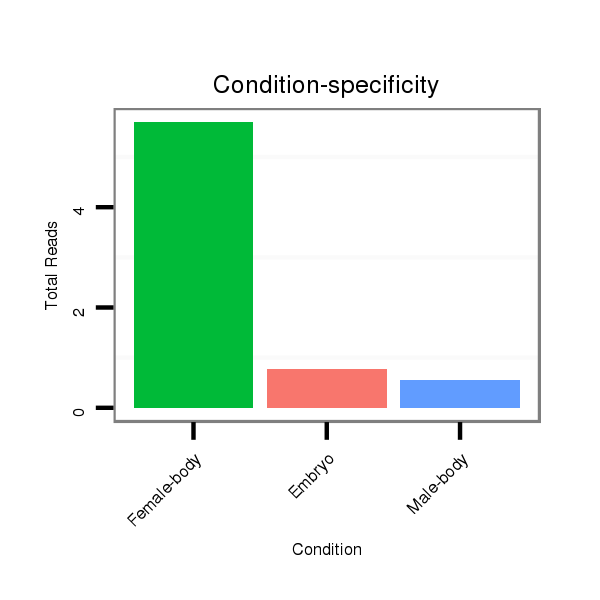
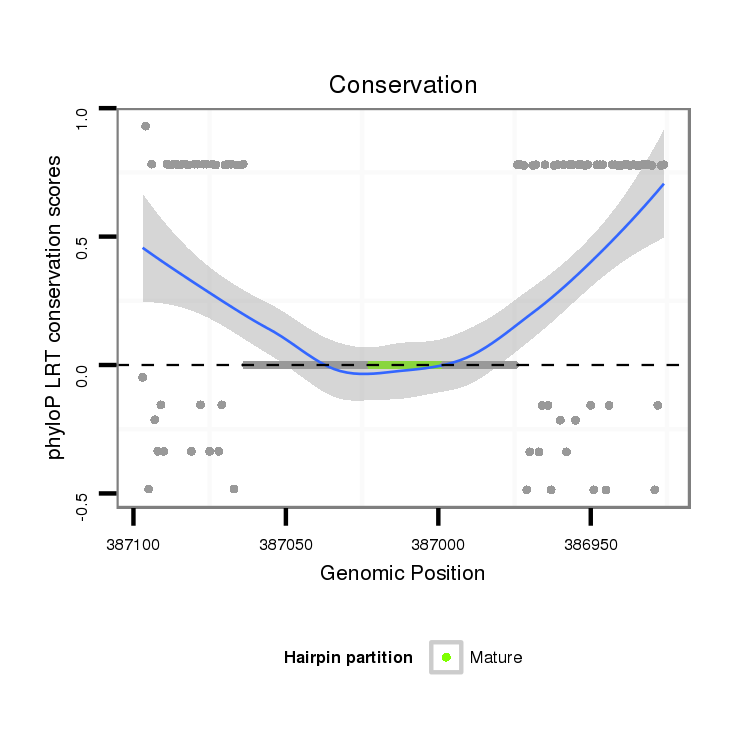
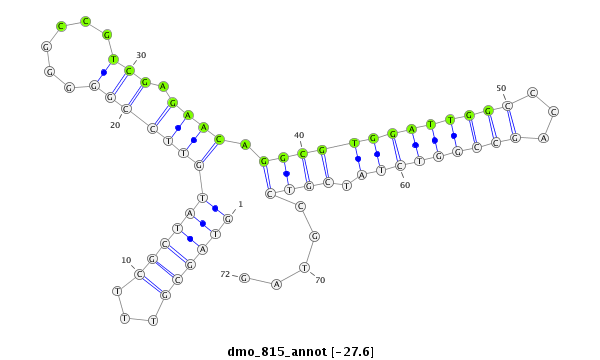
ID:dmo_815 |
Coordinate:scaffold_6482:386976-387047 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -27.6 |
 |
CDS [Dmoj\GI14383-cds]; exon [dmoj_GLEANR_14874:1]; exon [dmoj_GLEANR_14874:2]; CDS [Dmoj\GI14383-cds]; intron [Dmoj\GI14383-in]
No Repeatable elements found
| mature | star |
| ##################################################------------------------------------------------------------------------################################################## GCCGATTAGAGGACGATCGAGGCGATAACCGATGTCGCGACTCCGCGCGGGTAGCGTTTCGCTATGTTCCGGGGGCCGTCGAGAACAGGCGTGGATTGGCCCAGCCGGTCTATCGTCCGTAGCGCACGAGCCGACAGTCGGGTGCAGGCCGCCGGGATCGAGATGCCAGATC **************************************************.(((((...)))))((((((((......))).)))))(((((((((((((...))))))))).)))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
V110 male body |
M060 embryo |
V041 embryo |
|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................CCGTCGAGAACAGGCGTGGATTGG......................................................................... | 24 | 0 | 9 | 1.89 | 17 | 17 | 0 | 0 | 0 |
| ...........................................................................CCGTCGAGAACAGGCGTGGATTGGC........................................................................ | 25 | 0 | 9 | 1.44 | 13 | 11 | 1 | 1 | 0 |
| ...........................................................................CCGTCGAGAACAGGCGTGGATTG.......................................................................... | 23 | 0 | 9 | 1.44 | 13 | 11 | 2 | 0 | 0 |
| ...........................................................................CCGTCGAGAACAGGCGTGGATTGGCCC...................................................................... | 27 | 0 | 9 | 0.44 | 4 | 2 | 1 | 1 | 0 |
| ............................................................................CGTCGAGAACAGGCGTGGATTGG......................................................................... | 23 | 0 | 9 | 0.44 | 4 | 4 | 0 | 0 | 0 |
| ............................................................................CGTCGAGAACAGGCGTGGATTG.......................................................................... | 22 | 0 | 9 | 0.33 | 3 | 3 | 0 | 0 | 0 |
| ............................................................................................................................CACGAGCCGACAGTCGGGTGCAGC........................ | 24 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 |
| ............................................................................................................................CACGAGCCGACAGTCGGGTGCA.......................... | 22 | 0 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................................................ACGAGCCGACAGTCGGGTGCAGG........................ | 23 | 0 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 |
| ............................................................................................................................CACGAGCCGACAGTCGGGTGCAGG........................ | 24 | 0 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 |
| ...........................................................................CCGTCGAGAACAGGCGTGGATTGGCC....................................................................... | 26 | 0 | 9 | 0.22 | 2 | 0 | 1 | 1 | 0 |
| ......................................................................GGGGGCCGTCGAGAACAGGCG................................................................................. | 21 | 0 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 |
| ...........................................................................CCGTCGAGAACAGGCGTGGATTGGCT....................................................................... | 26 | 1 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................TCCGGGGGCCGTCGAGAACAGGCG................................................................................. | 24 | 0 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................CGTCGAGAACAGGCGTGGAT............................................................................ | 20 | 0 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................CGTCGAGAACAGGCGTGGATTGATA....................................................................... | 25 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 |
| GCCGATTAGAGGACGATCGAGGC..................................................................................................................................................... | 23 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 |
| ........................................................TTTCGCTATGTTCCGGGGGCC............................................................................................... | 21 | 0 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 |
| ...........................................................................CCGTCGAGAACAGGCGTGGATTGGTAT...................................................................... | 27 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................AGAACAGGCGTGGATTGGCC....................................................................... | 20 | 0 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 |
| .................................................................................AGAACAGGCGTGGAGTGGCC....................................................................... | 20 | 1 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 |
| .............................................................................GTCGAGAACAGGCGTGGATTGGC........................................................................ | 23 | 0 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 |
| .........................................................CTCGCTATGTTCCGGGGG................................................................................................. | 18 | 1 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| .......................................................GTTTCGCTATGTTCCGGGGGC................................................................................................ | 21 | 0 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 |
| ..........................................................TCGCTATGTTCCGGGGGC................................................................................................ | 18 | 0 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| ......................................................CGTTTCGCTATGTTCCGGGG.................................................................................................. | 20 | 0 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................GATGGAGGCCGCCGGGATAG............ | 20 | 3 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 |
|
CGGCTAATCTCCTGCTAGCTCCGCTATTGGCTACAGCGCTGAGGCGCGCCCATCGCAAAGCGATACAAGGCCCCCGGCAGCTCTTGTCCGCACCTAACCGGGTCGGCCAGATAGCAGGCATCGCGTGCTCGGCTGTCAGCCCACGTCCGGCGGCCCTAGCTCTACGGTCTAG
**************************************************.(((((...)))))((((((((......))).)))))(((((((((((((...))))))))).)))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V041 embryo |
V110 male body |
V056 head |
|---|---|---|---|---|---|---|---|---|---|---|
| .....AATCTCCTGCTAGCTCCGCTATTGGCT............................................................................................................................................ | 27 | 0 | 10 | 3.10 | 31 | 30 | 1 | 0 | 0 | 0 |
| ......ATCTCCTGCTAGCTCCGCTATTGGCT............................................................................................................................................ | 26 | 0 | 10 | 2.30 | 23 | 19 | 2 | 1 | 1 | 0 |
| .......TCTCCTGCTAGCTCCGCTATTGGCT............................................................................................................................................ | 25 | 0 | 10 | 2.20 | 22 | 15 | 7 | 0 | 0 | 0 |
| ............................................................CGATACAAGGCCCCCGGCAGCTCTT....................................................................................... | 25 | 0 | 10 | 1.90 | 19 | 6 | 10 | 3 | 0 | 0 |
| ..........................................................AGCGATACAAGGCCCCCGGCAGCTCTT....................................................................................... | 27 | 0 | 10 | 1.60 | 16 | 15 | 0 | 0 | 1 | 0 |
| ...........................................................GCGATACAAGGCCCCCGGCAGCTCTT....................................................................................... | 26 | 0 | 10 | 1.20 | 12 | 10 | 2 | 0 | 0 | 0 |
| ........CTCCTGCTAGCTCCGCTATTGGCT............................................................................................................................................ | 24 | 0 | 10 | 1.00 | 10 | 8 | 2 | 0 | 0 | 0 |
| ........................................................AAAGCGATACAAGGCCCCCGGCAGCTCTT....................................................................................... | 29 | 0 | 10 | 0.50 | 5 | 3 | 2 | 0 | 0 | 0 |
| ..........CCTGCTAGCTCCGCTATTGGCT............................................................................................................................................ | 22 | 0 | 10 | 0.40 | 4 | 1 | 1 | 0 | 2 | 0 |
| .........................................................AAGCGATACAAGGCCCCCGGCAGCTCTT....................................................................................... | 28 | 0 | 10 | 0.40 | 4 | 4 | 0 | 0 | 0 | 0 |
| .............................................................GATACAAGGCCCCCGGCAGCTCTT....................................................................................... | 24 | 0 | 10 | 0.30 | 3 | 3 | 0 | 0 | 0 | 0 |
| .................................................................CAAGGCCCCCGGCAGCTCTT....................................................................................... | 20 | 0 | 10 | 0.30 | 3 | 1 | 2 | 0 | 0 | 0 |
| ......................................................................CCCCCGGCAGCTCTTGTCCGCACCT............................................................................. | 25 | 0 | 8 | 0.25 | 2 | 0 | 2 | 0 | 0 | 0 |
| ........................................................................CCCGGCAGCTCTTGTCCGCACCT............................................................................. | 23 | 0 | 9 | 0.22 | 2 | 0 | 2 | 0 | 0 | 0 |
| ....TAATCTCCTGCTAGCTCCGCTATTGGCT............................................................................................................................................ | 28 | 0 | 10 | 0.20 | 2 | 2 | 0 | 0 | 0 | 0 |
| CGGCTAATCTCCTGCTAGCTCCGCTAT................................................................................................................................................. | 27 | 0 | 10 | 0.20 | 2 | 1 | 1 | 0 | 0 | 0 |
| .....AATCTCCTGCTAGCTCCGCTATTGGC............................................................................................................................................. | 26 | 0 | 10 | 0.20 | 2 | 2 | 0 | 0 | 0 | 0 |
| .........TCCTGCTAGCTCCGCTATTGGCTGCATC....................................................................................................................................... | 28 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................CAGCTCTTGTCCGCACCTAACCGGGT..................................................................... | 26 | 0 | 13 | 0.15 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................CCGGCGGCCCTAGCTCTACGG..... | 21 | 0 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................CCCCGGCAGCTCTTGTCCGCACCT............................................................................. | 24 | 0 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................CCACCTCCGTCTGCCCTAGCT........... | 21 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................CGGCAGCTCTTGTCCGCACCTAACCGGGT..................................................................... | 29 | 0 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................GCAGCTCTTGTCCGCACCTAACCGGTT..................................................................... | 27 | 1 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................TGCAGCTCTTGTCCGCACCTAACCGGGT..................................................................... | 28 | 1 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................GGCAGCTCTTGTCCGCACCTAACCGGGT..................................................................... | 28 | 0 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................GGCCCCCGGCAGCTCTTGT..................................................................................... | 19 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................CGATACAAGGCCCCCGGCAGCTCT........................................................................................ | 24 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................CGATACGAGGCCCCCGGCAGCTCTT....................................................................................... | 25 | 1 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| CGGCTAATCTCCTGCTAGCTC....................................................................................................................................................... | 21 | 0 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....TATCTCCTGCTAGCTCCGCTATTGGCT............................................................................................................................................ | 27 | 1 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......ATCTCCTGCTAGCTCCCCTATTGGCT............................................................................................................................................ | 26 | 1 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....AATCTCCTGCTAGCTCCGCTATTG............................................................................................................................................... | 24 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......ATCTCCTGCTAGCTCCGCTATTGGGT............................................................................................................................................ | 26 | 1 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| .GGCTAATCTCCTGCTAGCTCCGCTAT................................................................................................................................................. | 26 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................AGGCCCCCGGCAGCTCTT....................................................................................... | 18 | 0 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................CGATACAAGGCCCCCGGCAGCTATT....................................................................................... | 25 | 1 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................AGCGATACAAGGCCCCCGGCAGC........................................................................................... | 23 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................CGATACAAGGCCCCCTGCAGCTCTT....................................................................................... | 25 | 1 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................ACGATACAAGGCCCCCGGCAGCTCTTGT..................................................................................... | 28 | 1 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................GGCCCCCGGCAGCTCTTGTCC................................................................................... | 21 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| .GGCTAATCTCCTGCTAGCTCCGCTATT................................................................................................................................................ | 27 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................AGCGATACAAGGCCCCCGGCAGCTCTTG...................................................................................... | 28 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......CCTCCTGCTAGCTCCGCTATTGGCT............................................................................................................................................ | 25 | 1 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....GAATCTCCTGCTAGCTCCGCTATTGGCT............................................................................................................................................ | 28 | 1 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................ATACAAGGCCCCCGGCAGCTCTT....................................................................................... | 23 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....AGTCTCCTGCTAGCTCCGCTATTGGCT............................................................................................................................................ | 27 | 1 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........TTCCTGCTAGCTCCGCTATTGGCT............................................................................................................................................ | 24 | 1 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......ATCTCCTGCTAGCTCCGCTATT................................................................................................................................................ | 22 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................TACAAGGCCCCCGGCAGCTCTT....................................................................................... | 22 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................CGATACAAGGCCCCCGGCAGCTC......................................................................................... | 23 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...CTAATCTCCTGCTAGCTCCGCTATTGGCT............................................................................................................................................ | 29 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......ATCTCCTGCTAGCTCCGCTATTGGC............................................................................................................................................. | 25 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........TCCTGCTAGCTCCGCTATTGGCT............................................................................................................................................ | 23 | 0 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......CCTCCTGCTAGCTCCGCTATTGGGT............................................................................................................................................ | 25 | 2 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| CGGCTAATCTCCTGCTAGCTCCGCT................................................................................................................................................... | 25 | 0 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................AAAGCGATACAAGGCCCCCGGCAGCTCGT....................................................................................... | 29 | 1 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......ATCTCCTGCTAGCTCAGCTATTGGCT............................................................................................................................................ | 26 | 1 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...CTAATCTCCTGCTAGCTCCGCTATT................................................................................................................................................ | 25 | 0 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................CAAGGCCCCCGGCAGCTATT....................................................................................... | 20 | 1 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................CCGCACCTAACCGGGTCGGCCA............................................................... | 22 | 0 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................TAACCGGGTCGGCCAGATAGCAGGCGT................................................... | 27 | 1 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................ACCGCGTCGGCCATCTAGCAG....................................................... | 21 | 3 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................CCGGGTCGGCCAGATAGCAGGCGT................................................... | 24 | 1 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................CGCAAAGCGATACAAGGCCC................................................................................................... | 20 | 0 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................CCGGGTCGGCCAGATAGCA........................................................ | 19 | 0 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................TTGTCCGCACCCAAGCCGG...................................................................... | 19 | 3 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6482:386926-387097 - | dmo_815 | GCCGATTAGAGGACGATCGAGGCGATAACCGATGTCGCGACTCCGCGCGGGTAGCGTTTCGCTATGTTCCGGGGGCCGTCGAGAACAGGCGTGGATTGGCCCAGCCGGTCTATCGTCCGTAGCGCACGAGCCGACAGTCGGGTGCAGGCCGCCGGGATCGAGATGCCAGATC |
| droTak1 | scf7180000412386:5429-5430 - | GC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droYak3 | v2_chrUn_003:158478-158511 - | GCGGTGCCGAGGACGAGCGGGGAGAGGACCCATG------------------------------------------------------------------------------------------------------------------------------------------ | |
| droEre2 | scaffold_4845:2295159-2295208 - | C--------------------------------------------------------------------------------------------------------------------------GCAGTAGATGGGAGACTGGAGCAGAGCGCGAGGATCGAGATGCCACGTC |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 11:06 PM