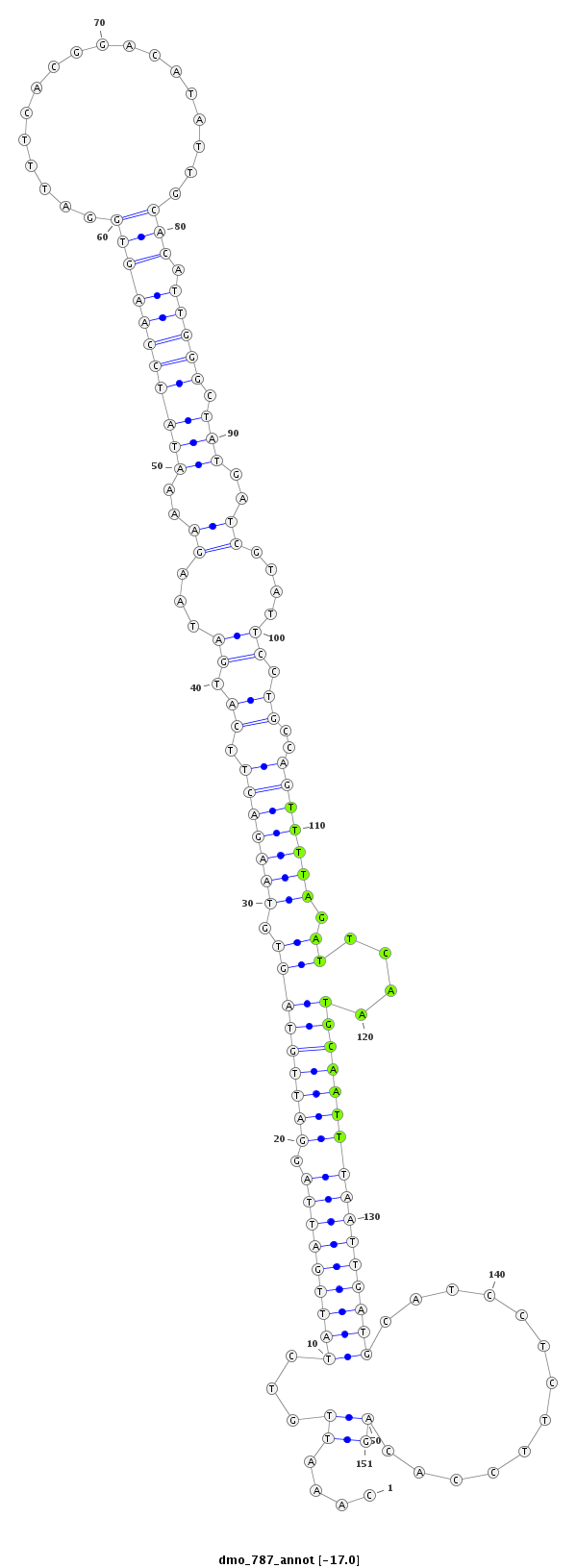
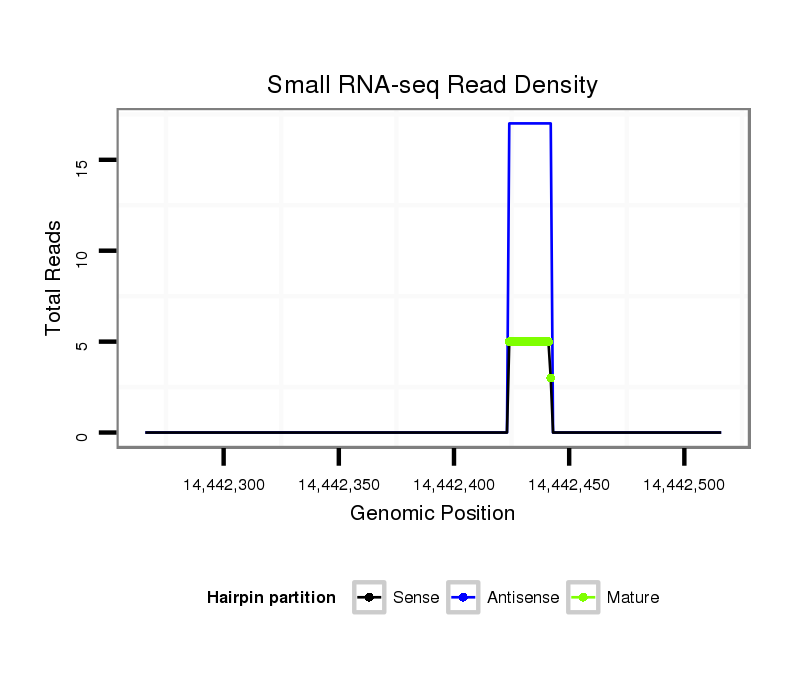
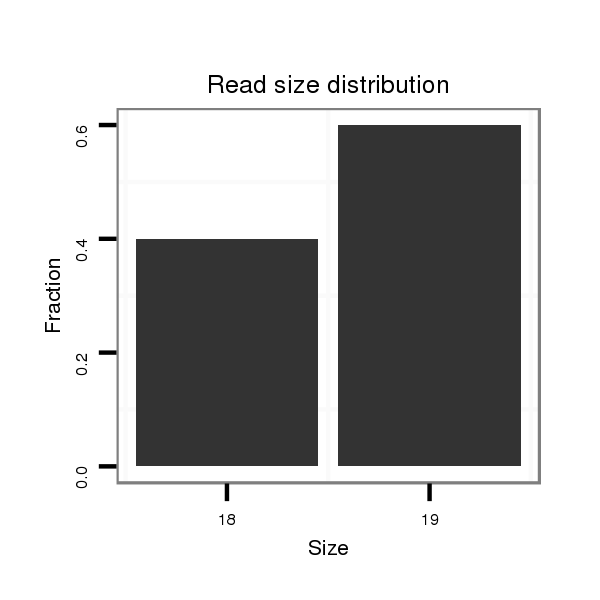


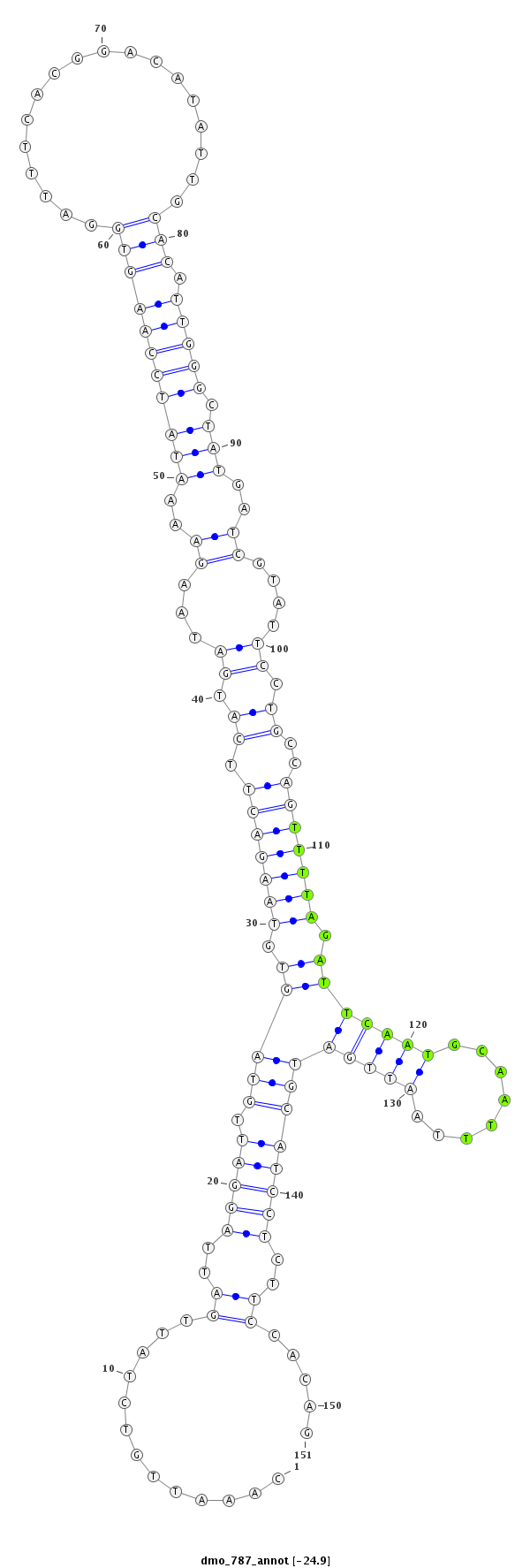
ID:dmo_787 |
Coordinate:scaffold_6473:14442316-14442466 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
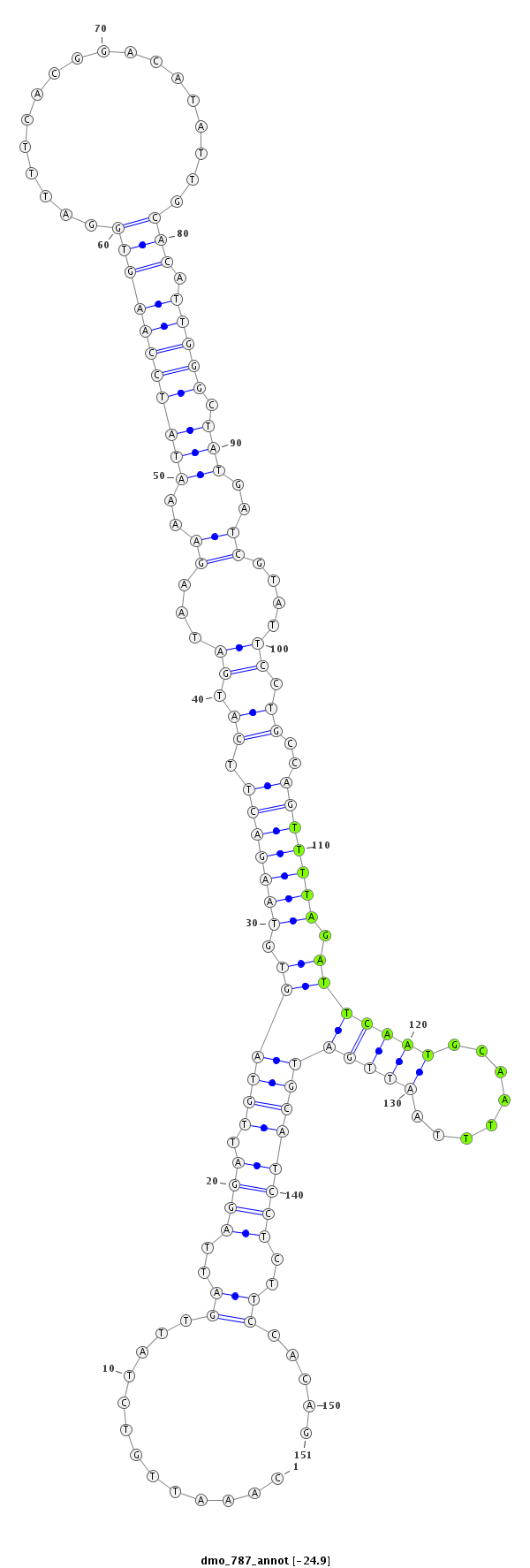
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -24.9 | -24.9 | -24.3 | -24.3 |
 |
 |
 |
 |
exon [dmoj_GLEANR_16629:4]; CDS [Dmoj\GI15928-cds]; intron [Dmoj\GI15928-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TCAGATCTATTCCCTATCAGTATGGACGCACTATAATATTAAATTGATTGCAAATTGTCTATTGATTAGGATTGTAGTGTAAGACTTCATGATAAGAAAATATCCAAGTGGATTTCACGGACATATTGCACATTGGGCTATGATCGTATTCCTGCCAGTTTTAGATTCAATGCAATTTAATTGATGCATCCTCTTCCACAGCTCAATTTAAGTGTGAATAAAAAGCGGAGCCAAACGGAAAATACATCGGA **************************************************....((...(((((((((.(((((((((.(((((((.((.((...((..(((((((((((..................))).))))).)))..))....)).))..))))))).))....)))))))))))))))).............))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
V110 male body |
V056 head |
|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................ATTTTTAGATTCAATGCAAT........................................................................... | 20 | 1 | 1 | 25.00 | 25 | 18 | 6 | 1 |
| ............................................................................................................................................................ATTTTTAGATTCAATGCAATT.......................................................................... | 21 | 1 | 1 | 12.00 | 12 | 11 | 1 | 0 |
| .............................................................................................................................................................TTTTTAGATTCAATGCAAT........................................................................... | 19 | 1 | 1 | 7.00 | 7 | 7 | 0 | 0 |
| ...........................................................................................................................................................TATTTTTAGATTCAATGCAAT........................................................................... | 21 | 2 | 2 | 5.00 | 10 | 8 | 1 | 1 |
| ..............................................................................................................................................................TTTTAGATTCAATGCAATT.......................................................................... | 19 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 |
| ..............................................................................................................................................................TTTTAGATTCAATGCAAT........................................................................... | 18 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| ............................................................................................................................................................ATTTTTAGATTCAATGCAA............................................................................ | 19 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| .............................................................................................................................................................TTTTTAGATTCAATGCAATT.......................................................................... | 20 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| ...........................................................................................................................................................TATTTTTAGATTCAATGCAATT.......................................................................... | 22 | 2 | 2 | 1.50 | 3 | 3 | 0 | 0 |
| ..............................................................................................................................................................TTTTAGATTCAATGCAATTA......................................................................... | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..............................................................................................................................................................TTTTAGATTCAATGCAATTG......................................................................... | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..............................................................................................................................................................TTTTAGATTCAATGCAATTAC........................................................................ | 21 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| ............................................................................................................................................................ATTTTTAGATTCAATGCAATTA......................................................................... | 22 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| ...........................................................................................................................................................TATTTTTAGATTCAATGCAA............................................................................ | 20 | 2 | 5 | 0.40 | 2 | 1 | 0 | 1 |
| ..........................................................................................................................................................TTATTTTTAGATTCAATGCAAT........................................................................... | 22 | 3 | 19 | 0.37 | 7 | 4 | 3 | 0 |
| ............................................................................................................................................................ATTTTTAGATTCAATGCAATTAC........................................................................ | 23 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 |
| ...............................................................................................................................................................TTTAGATTCAATGCAATTAC........................................................................ | 20 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 |
| ..........................................................................................................................................................TTATTTTTAGATTCAATGCAATT.......................................................................... | 23 | 3 | 12 | 0.17 | 2 | 2 | 0 | 0 |
| ...........................................................................................................................................................TATTTTTAGATTCAATGCA............................................................................. | 19 | 2 | 20 | 0.15 | 3 | 3 | 0 | 0 |
| ................................................................................................................................................................TTAGATTCAATGCAATTAC........................................................................ | 19 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 |
| ..........................................................................................................................................................TTATTTTTAGATTCAATGCAA............................................................................ | 21 | 3 | 20 | 0.10 | 2 | 2 | 0 | 0 |
| .....................................................ATTGTCTAGTGATTAGGA.................................................................................................................................................................................... | 18 | 1 | 12 | 0.08 | 1 | 1 | 0 | 0 |
|
AGTCTAGATAAGGGATAGTCATACCTGCGTGATATTATAATTTAACTAACGTTTAACAGATAACTAATCCTAACATCACATTCTGAAGTACTATTCTTTTATAGGTTCACCTAAAGTGCCTGTATAACGTGTAACCCGATACTAGCATAAGGACGGTCAAAATCTAAGTTACGTTAAATTAACTACGTAGGAGAAGGTGTCGAGTTAAATTCACACTTATTTTTCGCCTCGGTTTGCCTTTTATGTAGCCT
**************************************************....((...(((((((((.(((((((((.(((((((.((.((...((..(((((((((((..................))).))))).)))..))....)).))..))))))).))....)))))))))))))))).............))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
V110 male body |
V056 head |
M060 embryo |
|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................AAAATCTAAGTTACGTTAA.......................................................................... | 19 | 0 | 1 | 17.00 | 17 | 15 | 2 | 0 | 0 |
| ............................................................................................................................................................TAAAAATCTAAGTTACGTTAA.......................................................................... | 21 | 1 | 1 | 13.00 | 13 | 7 | 5 | 1 | 0 |
| ............................................................................................................................................................TAAAAATCTAAGTTACGTTA........................................................................... | 20 | 1 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 |
| ...........................................................................................................................................................ATAAAAATCTAAGTTACGTTAA.......................................................................... | 22 | 2 | 2 | 3.50 | 7 | 6 | 1 | 0 | 0 |
| ..............................................................................................................................................................AAAATCTAAGTTACGTTAATG........................................................................ | 21 | 2 | 2 | 2.00 | 4 | 2 | 1 | 1 | 0 |
| ...........................................................................................................................................................ATAAAAATCTAAGTTACGTTA........................................................................... | 21 | 2 | 2 | 1.50 | 3 | 3 | 0 | 0 | 0 |
| ...............................................................................................................................................................AAATCTAAGTTACGTTAATG........................................................................ | 20 | 2 | 4 | 1.00 | 4 | 3 | 1 | 0 | 0 |
| ..............................................................................................................................................................AAAATCTAAGTTACGTTAAT......................................................................... | 20 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................AAAAATCTAAGTTACGTTA........................................................................... | 19 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................AAAAATCTAAGTTACGTTAA.......................................................................... | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................TAAAAATCTAAGTTACGTTAATG........................................................................ | 23 | 3 | 4 | 0.75 | 3 | 2 | 1 | 0 | 0 |
| .................................................................................................................................................CACAAGGACGGCCAAAAT........................................................................................ | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 |
| ..............................................................................................................................................................AAAATCTGAGTTACGTTAA.......................................................................... | 19 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................AATCTAAGTTACGTTAATG........................................................................ | 19 | 2 | 7 | 0.29 | 2 | 1 | 1 | 0 | 0 |
| ...............................................................................................................................................TGCATAGGGACGGGCAAAAT........................................................................................ | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................AATAAAAATCTAAGTTACGTTA........................................................................... | 22 | 3 | 19 | 0.11 | 2 | 2 | 0 | 0 | 0 |
| .................................................................................................................................................................ATCTAAGTTACGTTAATG........................................................................ | 18 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................AATAAAAATCTAAGTTACGTT............................................................................ | 21 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6473:14442266-14442516 + | dmo_787 | TCAGATCTATTCCCTATCAGTATGGACGCACTATAATATTAAATTGATTGCAAATTGTCTATTGATTAGGATTGTAGTGTAAGACTTCATGATAAGAAAATATCCAAGTGGATTTCACGGACATATTGCACATTGGGCTATGATCGTATTCCTGCCAGTTTTAGATTCAATGCAATTTAATTGATGCATCCTCTTCCACAGCTCAATTTAAGTGTGAATAAAAAGCGGAGCCAAACGG------AAAATACATCGGA |
| droVir3 | scaffold_12928:2670917-2670975 + | CAGCAT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GATTTGTCTTTGAACAAGAGTCACAACGAGTCGGAGCAGGAGGATATATCGGA | |
| droGri2 | scaffold_15110:6158464-6158475 - | -----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGATAAGTCGGG | |
| droWil2 | scf2_1100000002375:7929-7951 + | CCAGATC-ACCCATTTTCCGTATG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
Generated: 05/17/2015 at 11:02 PM