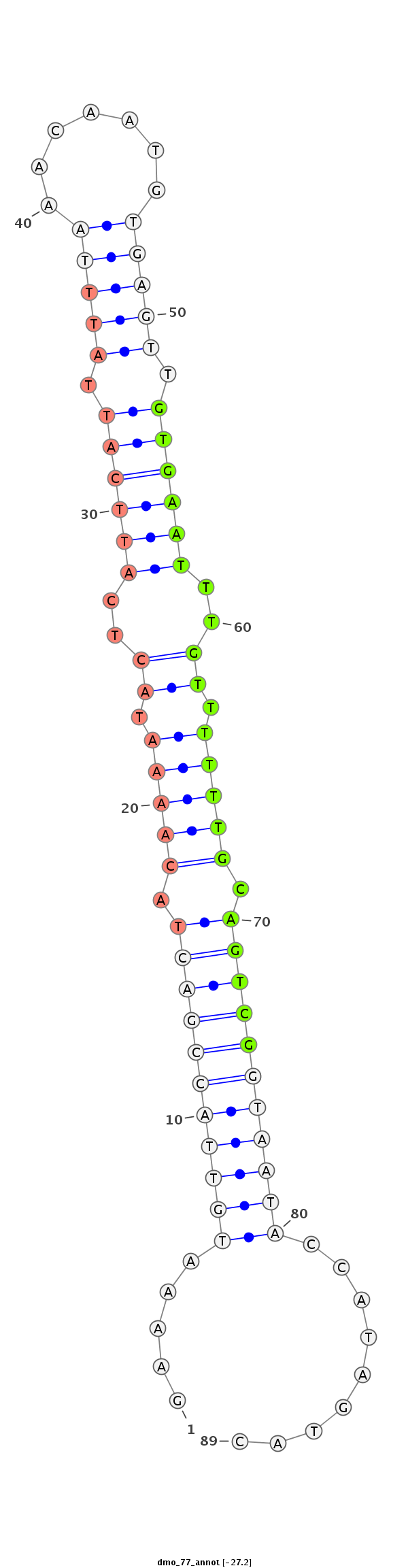
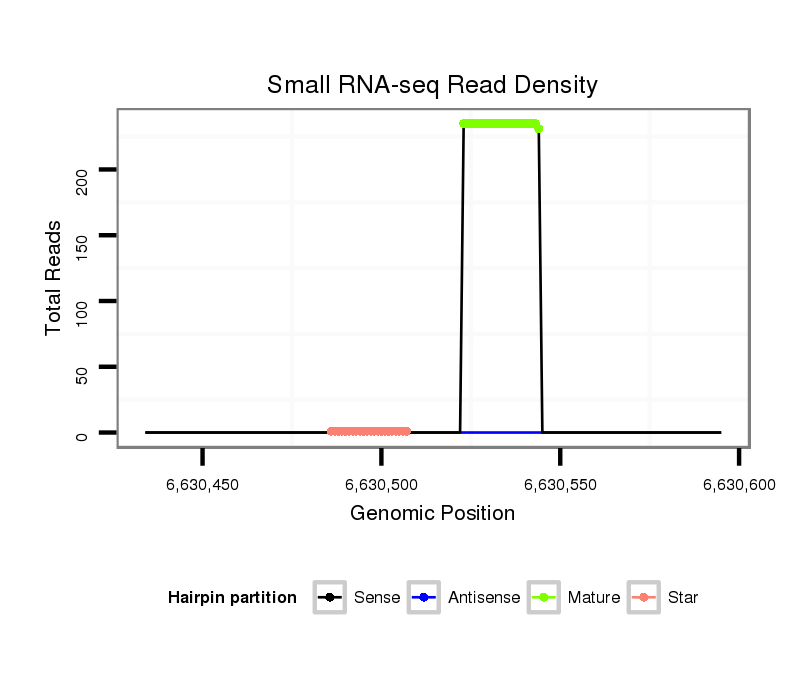
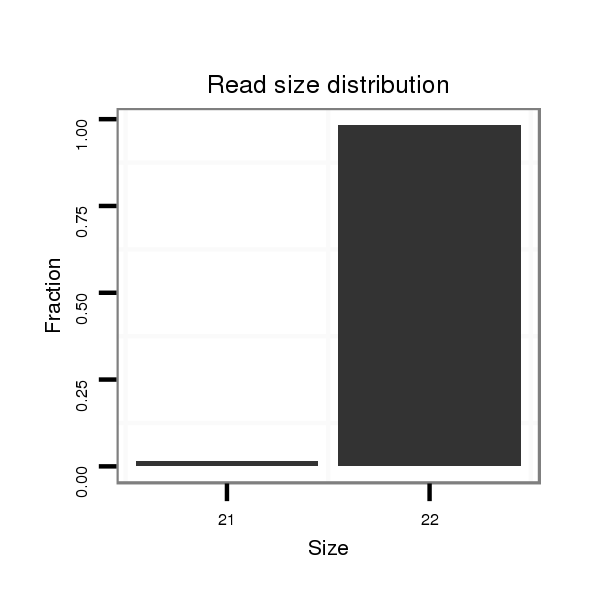
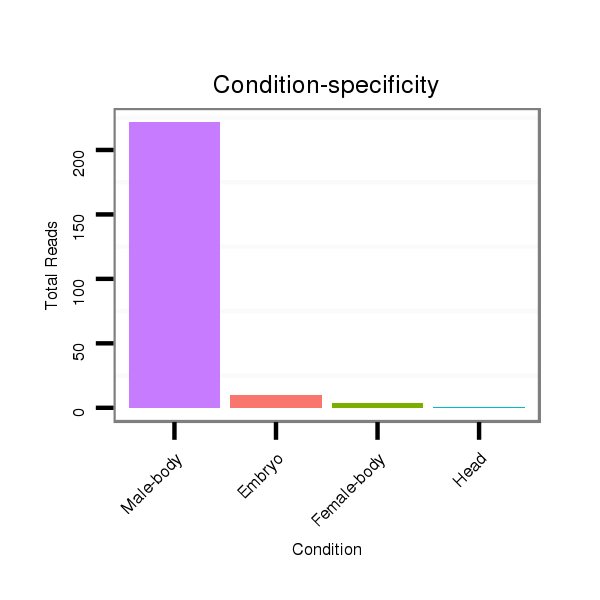
ID:dmo_77 |
Coordinate:scaffold_6540:6630484-6630545 + |
Confidence:candidate |
Class:Testes-restricted |
Genomic Locale:intergenic |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
intergenic
No Repeatable elements found
|
TATACATATATGTATATATACACCATTTGGCTCATTGGAAAATGTTACCGACTACAAAATACTCATTCATTATTTAAACAATGTGAGTTGTGAATTTGTTTTTTGCAGTCGGTAATACCATAGTACAACGCTCTTAATTGTTTTTGTTTGGATTTTATGTAT
*************************************.....(((((((((((.(((((.((..((((((.(((((.......))))).))))))..)).))))).))))))))))).........************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | V110 male body |
M060 embryo |
M046 female body |
V041 embryo |
V056 head |
|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................GTGAATTTGTTTTTTGCAGTCG................................................... | 22 | 0 | 1 | 231.00 | 231 | 217 | 5 | 3 | 5 | 1 |
| .........................................................................................GTGAATTTGTTTTTTGCAGTC.................................................... | 21 | 0 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 |
| .........................................................................................GTGAATTTGTTTTTTGCAGTCT................................................... | 22 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .........................................................................................GTGTATTTGTTTTTTGCAGTCG................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................GTGAATTTGTTTTTTCCAGTCG................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................GTGAATTTGTTTTTTGCAGTGG................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................GTGAATTTGTTTTTTGCAGTCGT.................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................GTTAATTTGTTTTTTGCAGTCG................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................GTGAATTTGTTATTTGCAGTCG................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................GGGTATTTGTTTTTTGCAGTCG................................................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................TACAAAATACTCATTCATTATT........................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................GTGACTTTGTTTTTTGCAGTCG................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................CTGAATTTGTTTTTTGCAGTCG................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................GAGAATTTGTTTTTTGCAGTCG................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................ACTACAAAATACTCATTCATTA.......................................................................................... | 22 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................TAATACCTTAATACAAAGCTC............................. | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 |
|
ATATGTATATACATATATATGTGGTAAACCGAGTAACCTTTTACAATGGCTGATGTTTTATGAGTAAGTAATAAATTTGTTACACTCAACACTTAAACAAAAAACGTCAGCCATTATGGTATCATGTTGCGAGAATTAACAAAAACAAACCTAAAATACATA
************************************.....(((((((((((.(((((.((..((((((.(((((.......))))).))))))..)).))))).))))))))))).........************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | V110 male body |
M046 female body |
|---|---|---|---|---|---|---|---|
| ....................................CCTCTTGCGATGGCTGATG........................................................................................................... | 19 | 3 | 10 | 0.10 | 1 | 1 | 0 |
| ........................................................................................................CGACAGCCATTATGGAATA....................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droMoj3 | scaffold_6540:6630434-6630595 + | dmo_77 | candidate | TATACATATATGTATATATACACCATTTGGCTCATTGGAAAATGTTACCGACTACAAAATACTCATTCATTATTTAAACAATGTGAGTTGTGAATTTGTTTTTTGCAGTCGGTAATACCATAGTACAACGCTCTTAATTGTTTTTGTTTGGATTTTATGTAT |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
Generated: 10/20/2015 at 06:59 PM