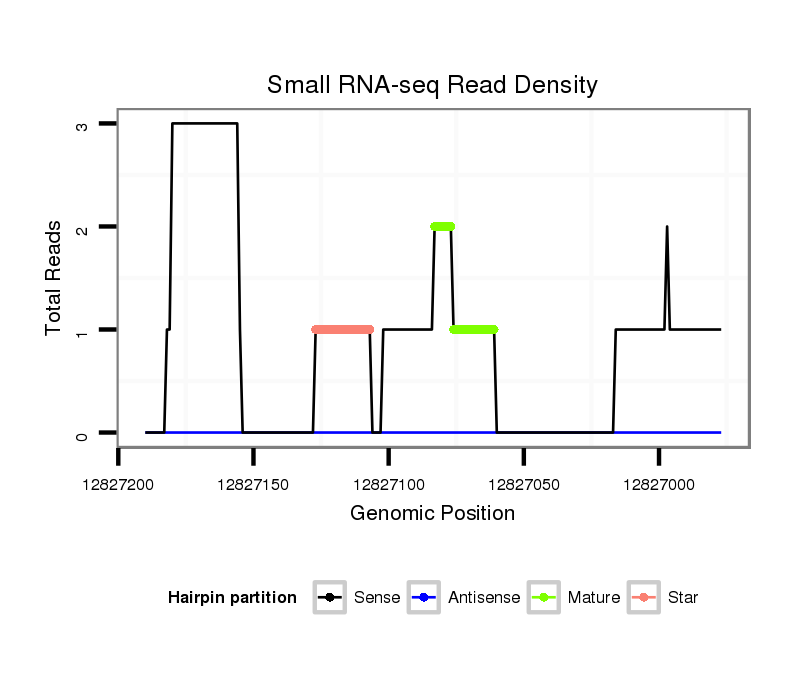
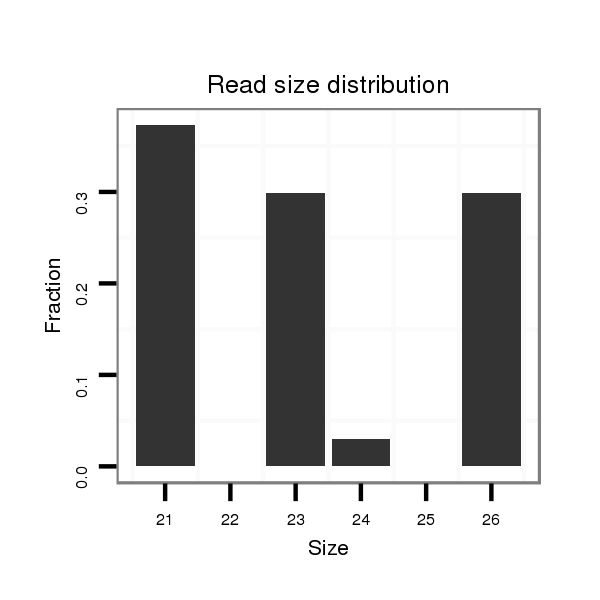
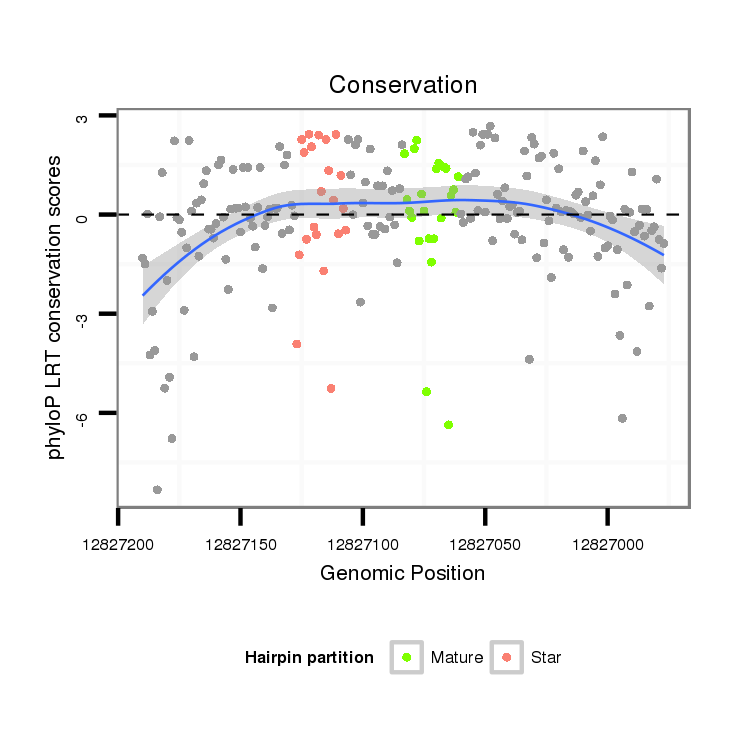
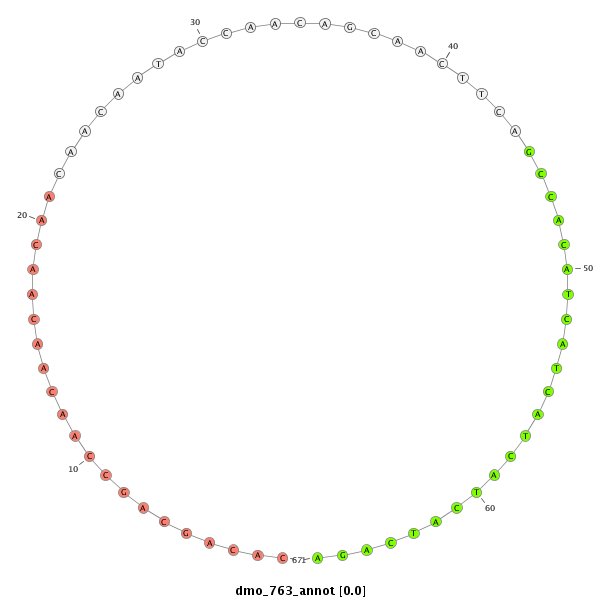
| GCAACATC-----AGCTC-------------------------------------------------------------------------CT-----------CCCG-----------CAGCAC------CACTT---------------------------------GCAACAT------------------------------------------------------CACCTGCAACATC-------------------------------------------------------ACCACCACCAACAA-----------------CAACAA---CAA---------CAGCAGC------AGC---AAC--------------AACAAC----A----G------------------CA------------A------------------CATC-------------------------------AGCTTT---------TGC----------CACAACAA---CAACAACAG------CA----------------------A---CAGCAG---CAG--------------------------CAAC----AA---------CAA------------------------------CAACAACATCATG-AC--------TTCCT---CAGCGCCGCTCTACT-----------------------ATC---CGCCCCCTCAT--CCCTCTC--CGGCG | Size | Hit Count | Total Norm | Total | M044
Female-body | M058
Embryo | V039
Embryo | V055
Head | V105
Male-body | V106
Head |
| ......................................................................................................................................................................................................................................................................................................................AA-----------------CAACAA---CAA---------CAGCAGC------AGC---.......................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AACAACAG------CA----------------------A---CAGCAG---CAG--------------------------CAAC----A.................................................................................................................................................. | 25 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................................................................................................................................................................................................................................................................................................................AACAA-----------------CAACAA---CAA---------CAGCAGC------A............................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................A----------------------A---CAGCAG---CAG--------------------------CAAC----AA---------CAA------------------------------C...................................................................................................... | 21 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................CAA-----------------CAACAA---CAA---------CAGCAGC------................................................................................................................................................................................................................................................................................................................................................................................................................ | 19 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAACAG------CA----------------------A---CAGCAG---CAG--------------------------CAAC----................................................................................................................................................... | 23 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................................................CAGCAGC------AGC---AAC--------------AACAAC----A----G------------------CA------------A------------------C..................................................................................................................................................................................................................................................................................................................... | 25 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................AA---------CAGCAGC------AGC---AAC--------------AACAAC----............................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AG------CA----------------------A---CAGCAG---CAG--------------------------CAAC----AA---------CAA------------------------------CA..................................................................................................... | 25 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AA---CAACAACAG------CA----------------------A---CAGCAG---C....................................................................................................................................................................................... | 21 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................AA---------CAGCAGC------AGC---AAC--------------AACAAC----A----G------------------C...................................................................................................................................................................................................................................................................................................................................................... | 24 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAA---CAACAACAG------CA----------------------A---CAGC............................................................................................................................................................................................. | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AACAG------CA----------------------A---CAGCAG---CAG--------------------------CAAC----................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................AA---CAA---------CAGCAGC------AGC---AAC--------------A........................................................................................................................................................................................................................................................................................................................................................................................ | 19 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAACAG------CA----------------------A---CAGCAG---CAG--------------------------C.......................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................CAA---------CAGCAGC------AGC---AAC--------------AACAAC----A----.......................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................A---------CAGCAGC------AGC---AAC--------------AACAAC----............................................................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................ACAA-----------------CAACAA---CAA---------CAGCAGC------AGC---.......................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................................................................................................................................................................................................................................GCAGC------AGC---AAC--------------AACAAC----A----G------------------C...................................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................................................................................................................................................................................................................................................................................................................AGCAGC------AGC---AAC--------------AACAAC----A----G------------------C...................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..............................................................................................................................................................................................................................................................................................................................................A---CAA---------CAGCAGC------AGC---AAC--------------A........................................................................................................................................................................................................................................................................................................................................................................................ | 18 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AACAG------CA----------------------A---CAGCAG---CAG--------------------------CAAC----A.................................................................................................................................................. | 22 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................CAA-----------------CAACAA---CAA---------CAGCAGC------AGC---A......................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................CAA---CAA---------CAGCAGC------AGC---AAC--------------A........................................................................................................................................................................................................................................................................................................................................................................................ | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AAC----AA---------CAA------------------------------CAACAACATCA............................................................................................ | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................A---CAGCAG---CAG--------------------------CAAC----AA---------CAA------------------------------C...................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................AACAA-----------------CAACAA---CAA---------CAGCAGC------AGC---.......................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAG---CAG--------------------------CAAC----AA---------CAA------------------------------CAACAAC................................................................................................ | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................A----------------------A---CAGCAG---CAG--------------------------CAAC----AA---------CA...................................................................................................................................... | 19 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................ACAA---CAA---------CAGCAGC------AGC---AAC--------------A........................................................................................................................................................................................................................................................................................................................................................................................ | 21 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................AACAA---CAA---------CAGCAGC------AGC---AA........................................................................................................................................................................................................................................................................................................................................................................................................ | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................AACAA-----------------CAACAA---CAA---------CAGCAGC------................................................................................................................................................................................................................................................................................................................................................................................................................ | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CA----------------------A---CAGCAG---CAG--------------------------CAAC----AA---------CAA------------------------------CA..................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAGCAG---CAG--------------------------CAAC----AA---------CAA------------------------------CAACA.................................................................................................. | 23 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAG------CA----------------------A---CAGCAG---CAG--------------------------CAAC----................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................GC------AGC---AAC--------------AACAAC----A----G------------------CA------------......................................................................................................................................................................................................................................................................................................................................... | 18 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |