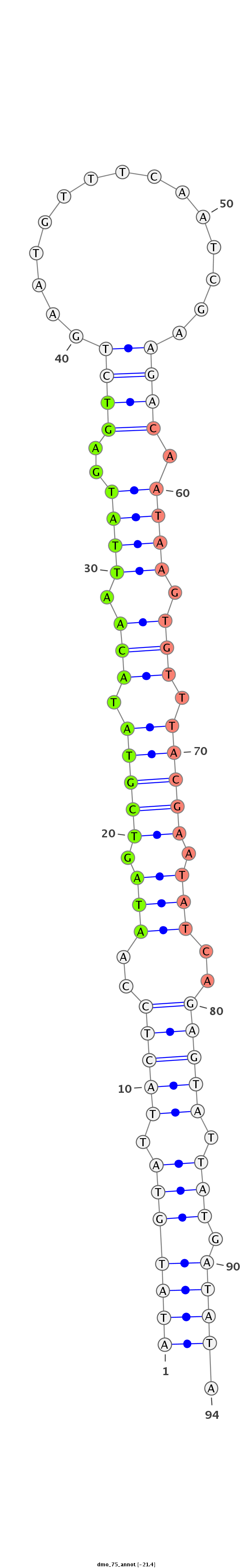
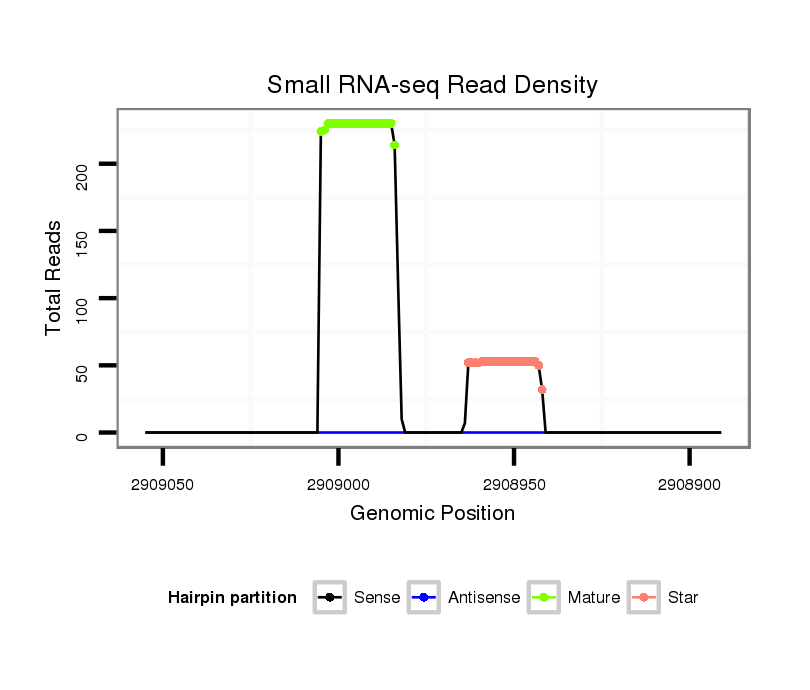

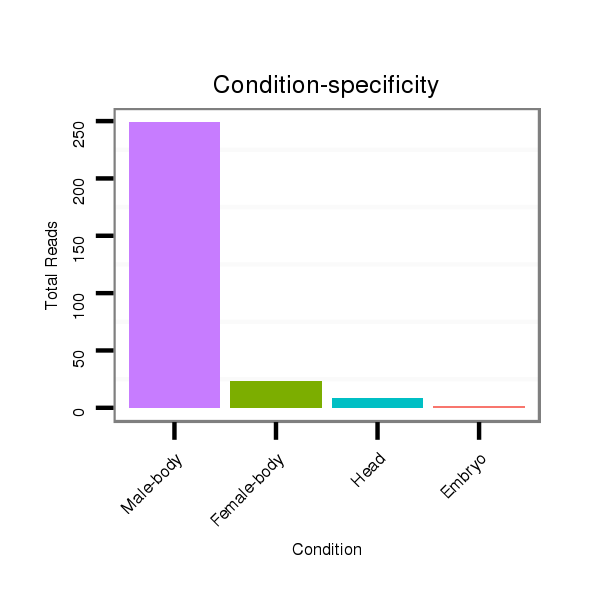
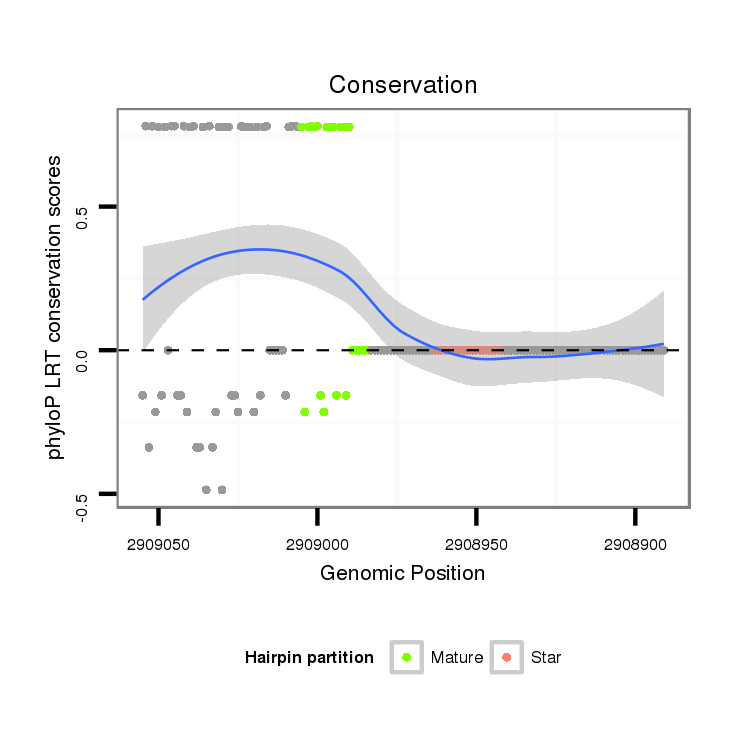
ID:dmo_75 |
Coordinate:scaffold_6328:2908941-2909005 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -21.1 | -20.7 | -20.6 |
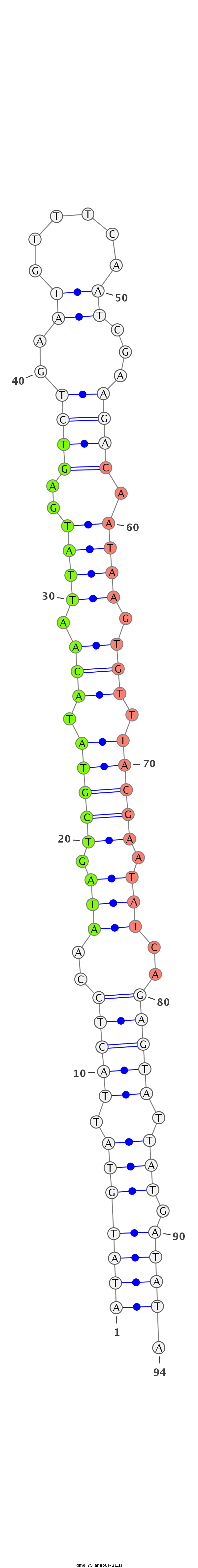 |
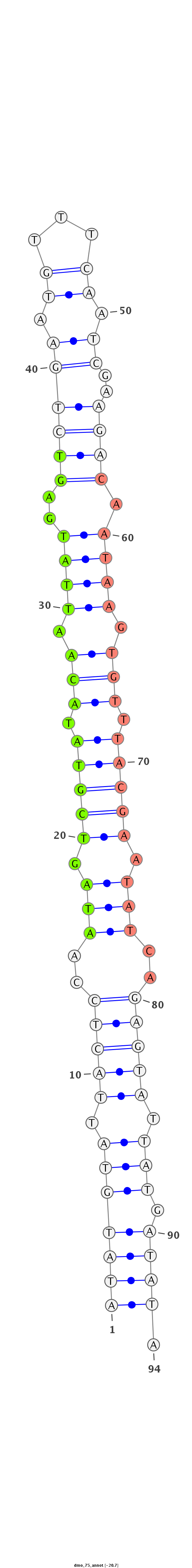 |
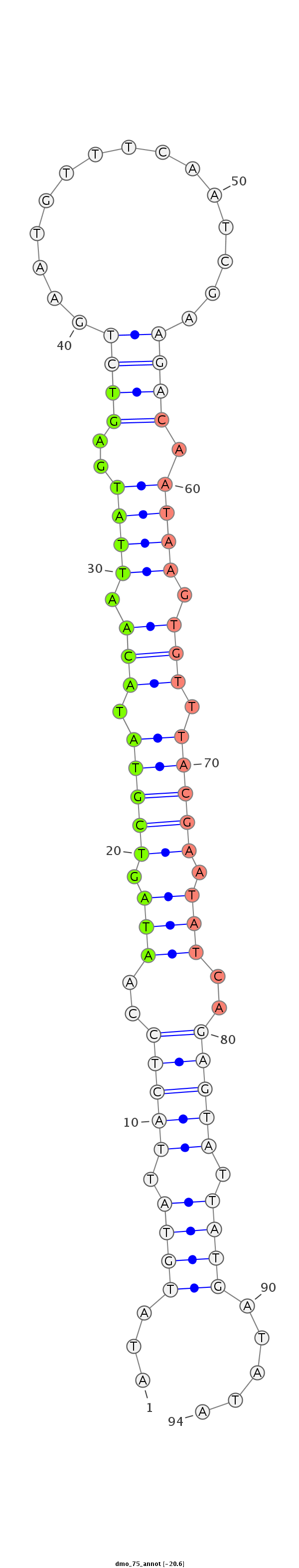 |
intergenic
No Repeatable elements found
|
AGTCTTATTCCCTCTAGATACCCTACAAACTGTATATATGTATTACTCCAATAGTCGTATACAATTATGAGTCTGAATGTTTCAATCGAAGACAATAAGTGTTTACGAATATCAGAGTATTATGATATAAGCACATACGGGAAAAGGATTATAGAGTGGCGACCC
***********************************(((((((.(((((..(((.(((((.(((.((((..((((...............)))).)))).))).))))).)))..))))).))).)))).************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | V110 male body |
M046 female body |
V056 head |
V049 head |
M060 embryo |
V041 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................ATAGTCGTATACAATTATGAGT............................................................................................. | 22 | 0 | 1 | 104.00 | 104 | 93 | 7 | 2 | 0 | 2 | 0 |
| ..................................................ATAGTCGTATACAATTATGAGTC............................................................................................ | 23 | 0 | 1 | 99.00 | 99 | 95 | 2 | 2 | 0 | 0 | 0 |
| ............................................................................................CAATAAGTGTTTACGAATATCA................................................... | 22 | 0 | 1 | 27.00 | 27 | 20 | 7 | 0 | 0 | 0 | 0 |
| ............................................................................................CAATAAGTGTTTACGAATATC.................................................... | 21 | 0 | 1 | 17.00 | 17 | 14 | 1 | 1 | 1 | 0 | 0 |
| ..................................................ATAGTCGTATACAATTATGAG.............................................................................................. | 21 | 0 | 1 | 16.00 | 16 | 15 | 0 | 1 | 0 | 0 | 0 |
| ....................................................AGTCGTATACAATTATGAGTCT........................................................................................... | 22 | 0 | 1 | 5.00 | 5 | 3 | 1 | 1 | 0 | 0 | 0 |
| ..................................................ATAGTCGTATACAATTATGAGTCT........................................................................................... | 24 | 0 | 1 | 5.00 | 5 | 4 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................ACAATAAGTGTTTACGAATATCA................................................... | 23 | 0 | 1 | 4.00 | 4 | 3 | 1 | 0 | 0 | 0 | 0 |
| ..................................................ATAGTCGTATACAATTATGAGTCC........................................................................................... | 24 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................ACAATAAGTGTTTACGAATAT..................................................... | 21 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ..................................................ATAGTCGTATACAATTATGAGC............................................................................................. | 22 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ATAGTCGTATACAATTATGAGTCTC.......................................................................................... | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ATAGTCGTATACAATTATGAGTT............................................................................................ | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................ATAGTCGTATAGAATTATGAGT............................................................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ATAGTCTTATACAATTATGAGT............................................................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ATAGTCATATACAATTATGAGT............................................................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................ATTCGTATGCAATTACGAGTCT........................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................CAATAAGTGTTTACGAATATT.................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................ACAATAAGTGTTTACGAATATC.................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................AAGTGTTTACGAATATCA................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................ACAATAAGTGTTTACGAATATCT................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................CAATAAGTGTTTACGAATATCC................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TAGTCGTATACAATTATGAGTCC........................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................ATAGTCGTATCCAATTATTAGT............................................................................................. | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................CAATAAGTGTTTACGGATATCA................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ATAGTCGTATCCAATTCTGAGTC............................................................................................ | 23 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................CAATAAGTGTTTACGAATAT..................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................CAATAAGTGTTTGCGAATATC.................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TAGTCGTATACAATTATGAGT............................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................CAATAAGTGTTTACGAATATCAAAAA............................................... | 26 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................ACATTCGGGATAAGGATT............... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................AACTACGGGAATAGGATT............... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................................................................................TCGGGAAAAGGATTGTGGA.......... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................GAATGTTTTAATCGACCACA....................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
|
TCAGAATAAGGGAGATCTATGGGATGTTTGACATATATACATAATGAGGTTATCAGCATATGTTAATACTCAGACTTACAAAGTTAGCTTCTGTTATTCACAAATGCTTATAGTCTCATAATACTATATTCGTGTATGCCCTTTTCCTAATATCTCACCGCTGGG
************************************(((((((.(((((..(((.(((((.(((.((((..((((...............)))).)))).))).))))).)))..))))).))).)))).*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V110 male body |
V041 embryo |
V056 head |
|---|---|---|---|---|---|---|---|---|
| ......TAAGGTAGACCTAGGGGATGT.......................................................................................................................................... | 21 | 3 | 2 | 1.00 | 2 | 1 | 1 | 0 |
| .....................................TACGTAATGAGGGCATCAG............................................................................................................. | 19 | 3 | 8 | 0.13 | 1 | 0 | 0 | 1 |
| .....................................TAGATAATGAGGGCATCAG............................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6328:2908891-2909055 - | dmo_75 | AGTCTTATTCCCTCTAGATACCCTACAAACTGTATATATGTATTACTCCAATAGTCGTATACAATTATGAGTCTGAATGTTTCAATCGAAGACAATAAGTGTTTACGAATATCAGAGTATTATGATATAAGCACATACGGGAAAAGGATTATAGAGTGGCGACCC |
| droEre2 | scaffold_4690:9682463-9682522 + | GGGCATGT-CCTCCAAGCGAGCAAAGAAGTAGTATTTGTG-----TTCCAAAAGTCAAATATAACT--------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/15/2015 at 02:24 PM