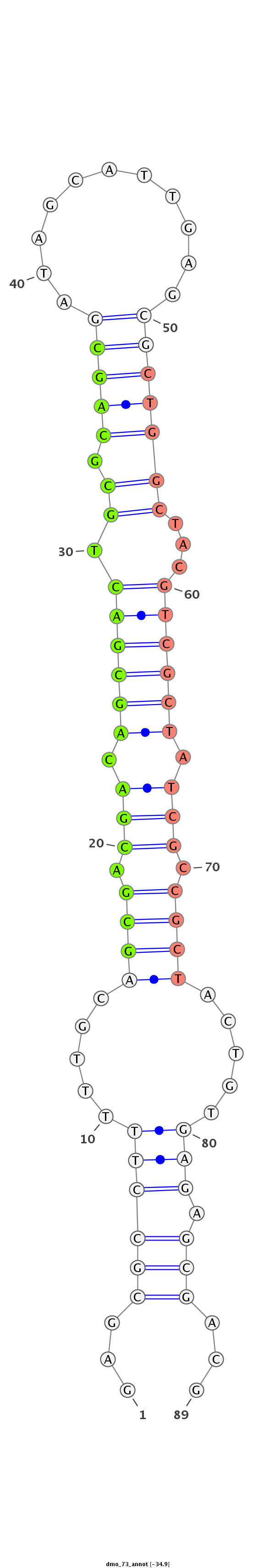
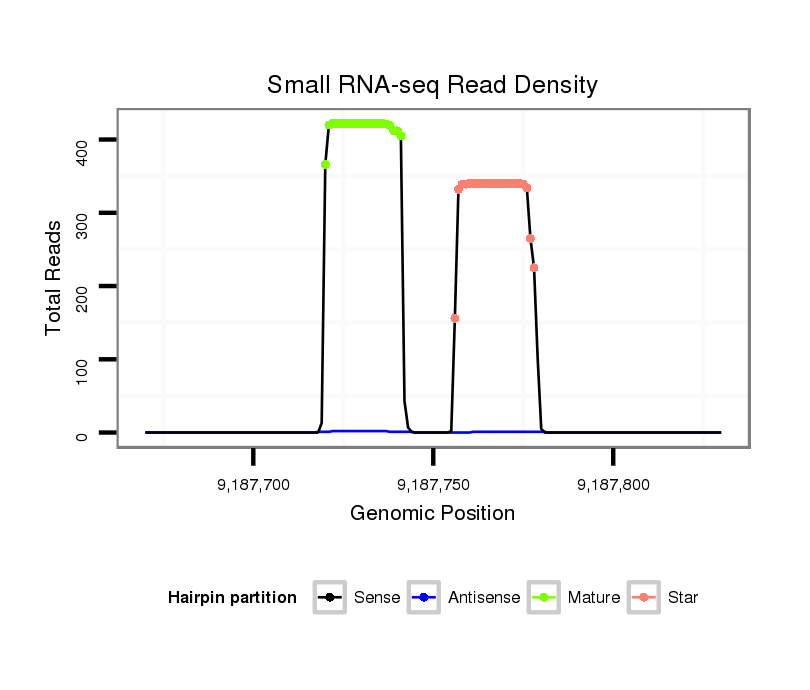
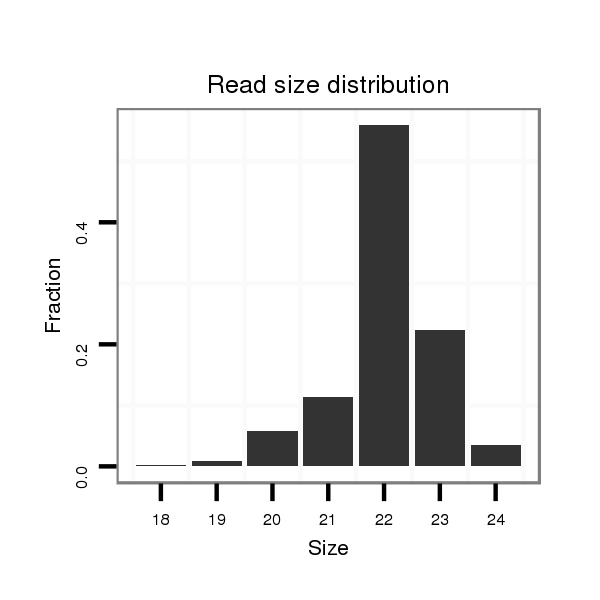
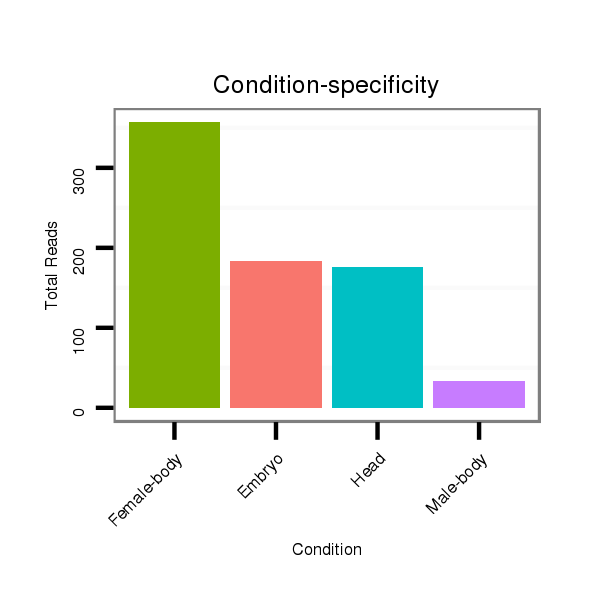
| ..................................................GCGACGACAGCGACTGCGCAGC......................................................................................... |
22 |
0 |
1 |
326.00 |
326 |
160 |
124 |
14 |
14 |
6 |
8 |
| ......................................................................................CTGGCTACGTCGCTATCGCCGCT.................................................... |
23 |
0 |
1 |
76.00 |
76 |
40 |
8 |
12 |
6 |
8 |
2 |
| .......................................................................................TGGCTACGTCGCTATCGCCGCTA................................................... |
23 |
0 |
1 |
73.00 |
73 |
28 |
4 |
21 |
15 |
3 |
2 |
| .......................................................................................TGGCTACGTCGCTATCGCCGCT.................................................... |
22 |
0 |
1 |
44.00 |
44 |
24 |
9 |
2 |
7 |
2 |
0 |
| .......................................................................................TGGCTACGTCGCTATCGCCG...................................................... |
20 |
0 |
1 |
34.00 |
34 |
4 |
0 |
12 |
14 |
4 |
0 |
| ......................................................................................CTGGCTACGTCGCTATCGCCG...................................................... |
21 |
0 |
1 |
34.00 |
34 |
9 |
1 |
9 |
12 |
2 |
1 |
| ...................................................CGACGACAGCGACTGCGCAGC......................................................................................... |
21 |
0 |
1 |
24.00 |
24 |
16 |
5 |
1 |
2 |
0 |
0 |
| ...................................................CGACGACAGCGACTGCGCAGCG........................................................................................ |
22 |
0 |
1 |
23.00 |
23 |
13 |
7 |
0 |
0 |
1 |
2 |
| ......................................................................................CTGGCTACGTCGCTATCGCCGC..................................................... |
22 |
0 |
1 |
20.00 |
20 |
13 |
1 |
3 |
3 |
0 |
0 |
| ......................................................................................CTGGCTACGTCGCTATCGCCGCTA................................................... |
24 |
0 |
1 |
19.00 |
19 |
6 |
1 |
4 |
4 |
4 |
0 |
| .......................................................................................TGGCTACGTCGCTATCGCCGC..................................................... |
21 |
0 |
1 |
19.00 |
19 |
10 |
2 |
3 |
4 |
0 |
0 |
| ..................................................GCGACGACAGCGACTGCGCAGCG........................................................................................ |
23 |
0 |
1 |
13.00 |
13 |
10 |
1 |
0 |
1 |
1 |
0 |
| .................................................AGCGACGACAGCGACTGCGCAGC......................................................................................... |
23 |
0 |
1 |
11.00 |
11 |
3 |
6 |
1 |
0 |
0 |
1 |
| ..................................................GGGACGACAGCGACTGCGCAGC......................................................................................... |
22 |
1 |
1 |
10.00 |
10 |
0 |
10 |
0 |
0 |
0 |
0 |
| ......................................................................................CTGGCTACGTCGCTGTCGCCGCT.................................................... |
23 |
1 |
1 |
7.00 |
7 |
1 |
0 |
3 |
3 |
0 |
0 |
| ..................................................GCGACGACAGCGACTGCGC............................................................................................ |
19 |
0 |
1 |
7.00 |
7 |
7 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................CTGGCTACGTCGCTATCGCCGCC.................................................... |
23 |
1 |
1 |
6.00 |
6 |
0 |
6 |
0 |
0 |
0 |
0 |
| ......................................................................................CTGGCTACGTCGCTATCGCCGCTT................................................... |
24 |
1 |
1 |
6.00 |
6 |
4 |
1 |
0 |
1 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCTATCGCCGCTAC.................................................. |
24 |
0 |
1 |
5.00 |
5 |
0 |
2 |
2 |
1 |
0 |
0 |
| ..................................................GCGACGACAGCGACTGCGCAGCGT....................................................................................... |
24 |
1 |
1 |
5.00 |
5 |
0 |
0 |
1 |
0 |
4 |
0 |
| ..................................................GCGACGACAGCGACTGCGCAG.......................................................................................... |
21 |
0 |
1 |
5.00 |
5 |
3 |
0 |
0 |
0 |
2 |
0 |
| ......................................................................................CTGGCTACGTCGCTATCGCC....................................................... |
20 |
0 |
1 |
5.00 |
5 |
1 |
0 |
2 |
2 |
0 |
0 |
| ..................................................GCGTCGACAGCGACTGCGCAGC......................................................................................... |
22 |
1 |
1 |
4.00 |
4 |
0 |
3 |
1 |
0 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCTGTCGCCGCTA................................................... |
23 |
1 |
1 |
4.00 |
4 |
0 |
0 |
4 |
0 |
0 |
0 |
| ........................................................................................GGCTACGTCGCTATCGCCGCTA................................................... |
22 |
0 |
1 |
4.00 |
4 |
4 |
0 |
0 |
0 |
0 |
0 |
| ..................................................GTGACGACAGCGACTGCGCAGC......................................................................................... |
22 |
1 |
1 |
4.00 |
4 |
0 |
4 |
0 |
0 |
0 |
0 |
| ...................................................CGACGACAGCGACTGCGCAGCGA....................................................................................... |
23 |
0 |
1 |
4.00 |
4 |
2 |
1 |
0 |
1 |
0 |
0 |
| ...................................................CGACGACAGCGACTGCGCAGCGT....................................................................................... |
23 |
1 |
1 |
3.00 |
3 |
0 |
0 |
0 |
0 |
3 |
0 |
| .......................................................................................TGGCTACGTCGCTATCGCCGCC.................................................... |
22 |
1 |
1 |
3.00 |
3 |
0 |
3 |
0 |
0 |
0 |
0 |
| ......................................................................................CTGGCTACGTCGCTGTCGCCG...................................................... |
21 |
1 |
1 |
3.00 |
3 |
0 |
0 |
3 |
0 |
0 |
0 |
| ........................................................................................GGCTACGTCGCTATCGCCGCT.................................................... |
21 |
0 |
1 |
3.00 |
3 |
2 |
0 |
1 |
0 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCTATCGCCGCTT................................................... |
23 |
1 |
1 |
3.00 |
3 |
1 |
1 |
0 |
1 |
0 |
0 |
| ..................................................GCCACGACAGCGACTGCGCAGC......................................................................................... |
22 |
1 |
1 |
3.00 |
3 |
0 |
3 |
0 |
0 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCTGTCGCCG...................................................... |
20 |
1 |
2 |
2.50 |
5 |
1 |
0 |
1 |
3 |
0 |
0 |
| ......................................................................................CGGGCTACGTCGCTATCGCCGCT.................................................... |
23 |
1 |
1 |
2.00 |
2 |
0 |
2 |
0 |
0 |
0 |
0 |
| ......................................................................................CTGGCTACGTCGCTATCGCCGCTC................................................... |
24 |
1 |
1 |
2.00 |
2 |
0 |
0 |
1 |
0 |
1 |
0 |
| .......................................................................................TGGCTACGTCGCTATCGCCGCTAA.................................................. |
24 |
1 |
1 |
2.00 |
2 |
1 |
0 |
0 |
1 |
0 |
0 |
| ..................................................GCGACGACAGCGCCTGCGCAGC......................................................................................... |
22 |
1 |
1 |
2.00 |
2 |
0 |
1 |
0 |
1 |
0 |
0 |
| ...................................................CGACGACAGCGACTGCGCAG.......................................................................................... |
20 |
0 |
1 |
2.00 |
2 |
1 |
0 |
0 |
1 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCTATCTCCG...................................................... |
20 |
1 |
1 |
2.00 |
2 |
0 |
0 |
2 |
0 |
0 |
0 |
| ......................................................................................CTGGCTACGTCGCTGTCGCCGC..................................................... |
22 |
1 |
1 |
2.00 |
2 |
0 |
0 |
0 |
2 |
0 |
0 |
| ..................................................GCGACGACAGCGACTGCGCCGC......................................................................................... |
22 |
1 |
1 |
2.00 |
2 |
0 |
0 |
0 |
2 |
0 |
0 |
| ..................................................GCGACGACAGCGACTGCGCCGCGT....................................................................................... |
24 |
2 |
1 |
2.00 |
2 |
0 |
0 |
0 |
2 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCTATCGCCACTA................................................... |
23 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
| .......................................................................................TAGCTACGTCGCTATCGCCGCT.................................................... |
22 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
| ..................................................GCTACGACAGCGACTGCGCAGC......................................................................................... |
22 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
| ..........................................................................................CTACGTCGCTATCGCCGCTA................................................... |
20 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| ......................................................................................CTGGCTCCGCCGCTATCGTCGCT.................................................... |
23 |
3 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
| ..................................................GCGACGGCAGCGACTGCGCAGC......................................................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................TTGGCTACGTCGCTATCGCCGCT.................................................... |
23 |
1 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCTATCGCCGCTTA.................................................. |
24 |
2 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
| ......................................................................................CTGGCTACGTCGCTGTCGCCGCTT................................................... |
24 |
2 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
| .....................................................................................GCTGGCTACGTCGCTATCGCCG...................................................... |
22 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| ...................................................CTACGACAGCGACTGCGCAGCG........................................................................................ |
22 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCTATCGC........................................................ |
18 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCTATCGCCGCTATC................................................. |
25 |
2 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| ...................................................CGACGACAGCGACTGCGCCGC......................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
| .....................................................................................GCTGGCTACGTCGCTATCGCCGC..................................................... |
23 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..................................................GCGTCGACAGCGACTGCGCAGCT........................................................................................ |
23 |
2 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................CCTGGCTACGTCGCTATCGCCGCT.................................................... |
24 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................CTGGCTACGTCACTATCGCCGCTT................................................... |
24 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..................................................GCGACGACAGCGCCTGCGCAGA......................................................................................... |
22 |
2 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...........................................................................................TACGTCGCTGTCGCCGCTA................................................... |
19 |
1 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| ..................................................GCGACGACAGCGACTGCGCAGCGA....................................................................................... |
24 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................CTGGCTACGTCGCTATCGCCGCTTT.................................................. |
25 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..................................................GCGACGACAGCGAATGCGCAGC......................................................................................... |
22 |
1 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCTAGCGCCGTTA................................................... |
23 |
2 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| .......................................................................................TGGCTACGTCACTATCGCCGCTA................................................... |
23 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................CTGGCTACGTCGCTGTCGCCGCTA................................................... |
24 |
1 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| .......................................................................................TTGCTACGTCGCTATCGCCGCT.................................................... |
22 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...................................................CGACGACAGCGACTGTGCAGCGA....................................................................................... |
23 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................TGGCTACGTCGGTATCGCCGCTT................................................... |
23 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...................................................CGACGACAGCGACTGAGCAACGA....................................................................................... |
23 |
2 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCTATCGCCGCGT................................................... |
23 |
2 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCTATTGCCGCTA................................................... |
23 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ......................................................................................CGCGCTACGTCGCTATCGCCGCC.................................................... |
23 |
3 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCTATCCCCGCT.................................................... |
22 |
1 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| ..................................................GCGACGACGGCGACTGCGCAGC......................................................................................... |
22 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| .................................................AGCGACGACAGCGACTGCG............................................................................................. |
19 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCGATAGCAGCTA................................................... |
23 |
3 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| ................................................TCGCGACGACAGCGACTGCGCAGC......................................................................................... |
24 |
2 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| .................................................TGCGACGACAGCGACTGCGCAGC......................................................................................... |
23 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..................................................GCGACGACAGCGACTGCGCAGCT........................................................................................ |
23 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ......................................................................................CTGGCTACGTCTCTTTCTCCGCT.................................................... |
23 |
3 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| ..................................................GCGACGACAGCGACTGCGCAGCC........................................................................................ |
23 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ......................................................................................CGCGCTACGTCGCTATCGCCGCT.................................................... |
23 |
2 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ....................................................GACGACAGCGACTGCGCAGCGA....................................................................................... |
22 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...................................................CGACGACAGCGACTGCGCAGCGC....................................................................................... |
23 |
1 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCTATCGCCGCTAT.................................................. |
24 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...................................................CGACGACAGCGACTGCGCAGCGAT...................................................................................... |
24 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................TTGTTACGTCGCTATCGCCGCTA................................................... |
23 |
2 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGACGACAGCGACTGCGCGGC......................................................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................CTGGCTACGTCGCCGTCGCCGCT.................................................... |
23 |
2 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| ...................................................CGACGACAGCGACTGCTCAGC......................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
| ......................................................................................CTGGCTACGTCGCTGGCGCCG...................................................... |
21 |
2 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
| ......................................................................................CTGGCTACGGCGCTATCGCCG...................................................... |
21 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
| ......................................................................................CTGGCTACGTCGCTGTCGC........................................................ |
19 |
1 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| ......................................................................................CTGGCTACGTCCCTATCGCCGTT.................................................... |
23 |
2 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCTATCGCCTCTA................................................... |
23 |
1 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| ..................................................GCGACGACAGCGACTGCGCAGT......................................................................................... |
22 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ....................................................GACGACAGCGACTGCGCAGC......................................................................................... |
20 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
| ......................................................................................CTGGCTACGTCGCTATCGTCGCT.................................................... |
23 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
| ......................................................................................CTGGCTACGTCGCTATCGCCGTC.................................................... |
23 |
2 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
| ......................................................................................CTGGCTACGTCGCTATCGCCCCTC................................................... |
24 |
2 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| .......................................................................................TGGCCACGTCGCTATCGCCG...................................................... |
20 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCTATCGCCTCTC................................................... |
23 |
2 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| .................................................AGCTACGACAGCGACTGCGCAGC......................................................................................... |
23 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................TTGCTACGTCGCTATCGCCGCC.................................................... |
22 |
2 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGACTACAGCGACTGCGCAGC......................................................................................... |
22 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ......................................................................................CTGGCTACGTCGCTATCGTC....................................................... |
20 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
| ...................................................CGACGACAGCGACTGAGCAGC......................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| ......................................................................................CTGGCTACGTCGGTATCGCGG...................................................... |
21 |
2 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| ...................................................TGACGACAGCGACTGCGCAGCG........................................................................................ |
22 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................GCGGGCTACGGCGCTATCGTCG...................................................... |
22 |
3 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCTATCGCTGCTC................................................... |
23 |
2 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| .................................................ATCGACGACAGCGACTGCGCAGC......................................................................................... |
23 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCTCTCGCCGCT.................................................... |
22 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCTATCGCCGCTG................................................... |
23 |
1 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| .................................................AGCGACGACAGCGACTGC.............................................................................................. |
18 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ......................................................................................CTGGCTACGCCGCTATCGCCG...................................................... |
21 |
1 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| ......................................................................................CGGTCTACGTCGCTATCGCCGCC.................................................... |
23 |
3 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................TGGCTACGACGCTATCGCCGCTA................................................... |
23 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| .................................................AGCGACGACAGCGACTGCGCAGT......................................................................................... |
23 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ......................................................................................CTCGCTAAGTCGCTATCGCCGCC.................................................... |
23 |
3 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GCGACAACAGCGACTGCGCAGC......................................................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCTATCACCGCTA................................................... |
23 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCTCTCGCCGCTA................................................... |
23 |
1 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| ..................................................GCGACGACAGCGACTGCG............................................................................................. |
18 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................GACAACAACCATTTCGCCGAGT............. |
22 |
3 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
| ......................................................................................CGGGCGACGCCGCTATCGCCGCT.................................................... |
23 |
3 |
2 |
0.50 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCTGTCGCCGCTC................................................... |
23 |
2 |
2 |
0.50 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| ....................................................GACGACAGGGACTGCGCAGCTG....................................................................................... |
22 |
3 |
2 |
0.50 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| ..................................................GTGTCGACAGCGACTGCGCAGC......................................................................................... |
22 |
2 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................TGGCTACGTCGCTGTCGCCGCT.................................................... |
22 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................ATGGCTACGTCGCTGTCGCCGCT.................................................... |
23 |
2 |
2 |
0.50 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
| ..................................................GGGTCGACAGCGACTGCGCAGC......................................................................................... |
22 |
2 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................TGGCGACGGCGCTAGCGCCGCTA................................................... |
23 |
3 |
2 |
0.50 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| ..................................................GCGACGTCAGCGACCGCGCTGC......................................................................................... |
22 |
3 |
5 |
0.20 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| ...................................................CGACGACGGCGACTGCGCCGCG........................................................................................ |
22 |
2 |
6 |
0.17 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
| ......................................................................................CTGGCTACGTCCCGATCAC........................................................ |
19 |
3 |
14 |
0.07 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |