| droMoj3 |
scaffold_6473:1725400-1725770 + |
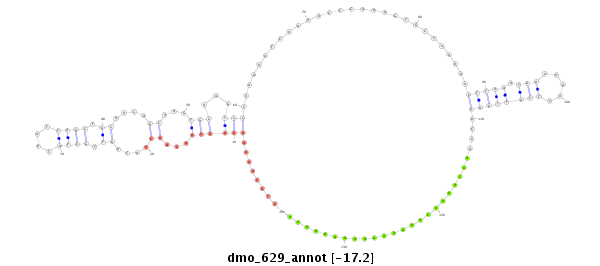
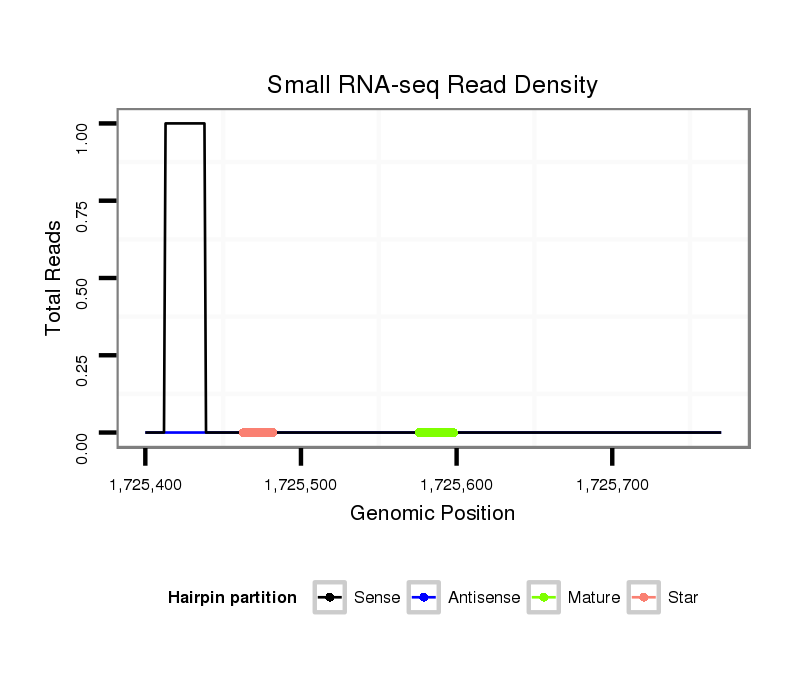
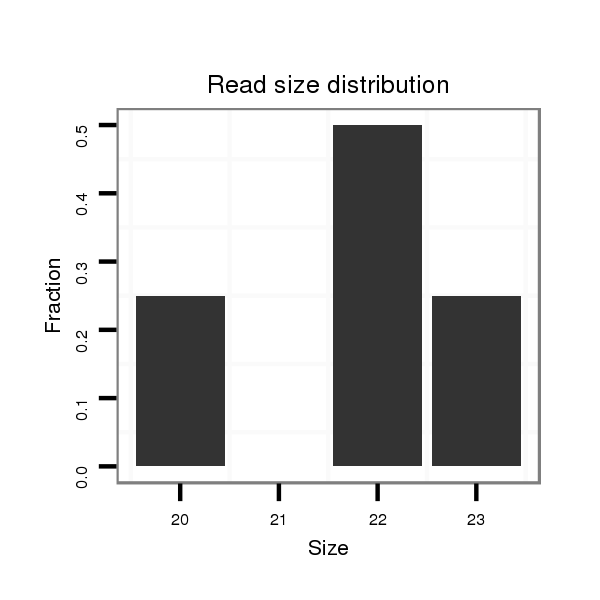
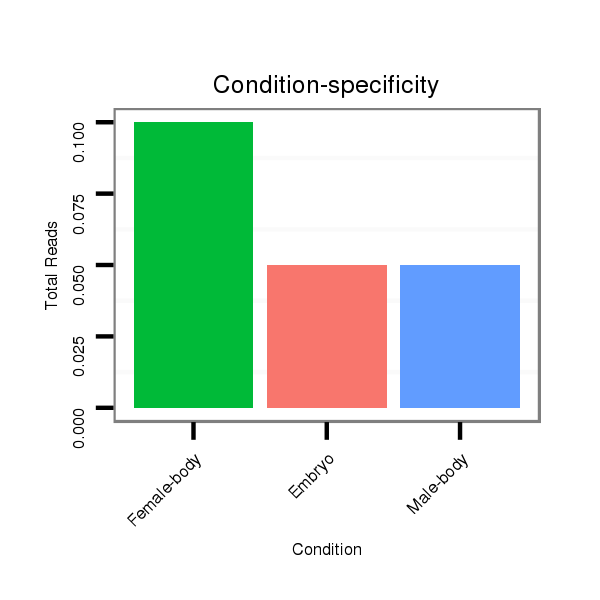
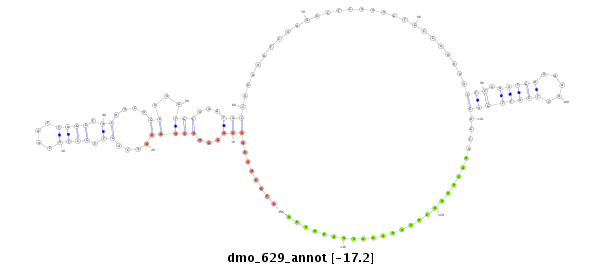
dmo_629 |
TTGA-ACGGTTGGACATCGAGGGCTTAAGCACGACCAACGTGAAGATGAAT----GTAAGTG------------------------C------------AGGAGCT--ACAACAGCAGCAGCAGCAACAGTTGCTATAATTA----------------GCTAC---AACA---------------GCAA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGCAAGTG-----------------------------CCAAAA------ATCAAAACTC--AAA--GTGCCAAAAACTCAAATGAA-----------------------AAACTCATTGAGAGCAAACAAAACAAAAAAAAAAAAAAAA------CAAAAAAAAAAGGACT--------AAA---------------------TTGTG-----TTTA-------------------------------------------------------------------------TTTA-------------------------------TAAATAATTTACTA------------TATTATCCAATTGGT----------------------------------------------TGGT-------------------------------------------------------------------------------------------------------------------------------------------TTTTTTTTTT---------AATGTGCT--C--AA--TTTT--TTTG---------T---TTC-----------------T----GTTTCTGTTGTG-----ATC----TT-TAAAGATAATTGGCGCTCTATTGGTGCTACT---CGCCTGCT--------------------------------------CGCTATGCGGCCTAAG |
| droVir3 |
scaffold_12928:1827355-1827686 + |
dvi_8533 |
TTGA-ACGATTGGACATCGAGGGCTCAAGCACGACCAACGTGAAGATGAAT----GTAAGTG------------------------C------------AGCAGCA--GCAGCAGCAGCAGCAGCAGC-------------------AGC----------------AGCA---------------ACAA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGCAAGTGATCAGAA--AAAAAAAAGAACAAAAAAACTGAA-ACTGAAAACGAAAA----------------------------------------------------------------------------------------------------CG-----AAAGGATT--------AAA---------------------ATGTG-----TT------------------A-------------------------------------------------------TGCAATATAAAAAATAGATTTGC----AATTTTTTTGCAATTA-------------------------------------------------------------------------CA----------------------------------------------------------------------------------------------TA------------------------GAATTCT-------------------TATTTTTTTTTG---------AATGTGC---T--AA--TTTT--GTTGTC-T----AT---TTT---------------TTTGTTTGTTT----TGTG-----GCC-------AAAAGTTGATTGGCGCTTTGTTAACGCTCGC---CGCCTGCT--------------------------------------CGCAATGTGGCTTAAT |
| droGri2 |
scaffold_15203:8768931-8769233 - |
|
TTTA-ACGGTTGGACATCGAGGGCTCCAGCACGACCAACGTGAAGATGAAT----GTAAGTG------------------------C------------AACAG--CAGCAGCACCACCAGCAGC----------------------AATAA---AATAATAATC-ATAA---------------AAAA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGCCAGT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GACA-ATATTCAAATTATAAATATATAA--CAAA----------------------------------------------------------------TATATTTATTTAGTATTTTGACG-------------------------CATATTTCAATTGATTTTTTTTTGTTAAC--------------------------------------------------------------------------------------------------------------------------------------------------------GTGC---------------------------------------------AACT--CCTT--TTGCT--CTACT----TT---TTT---------------TTTCTTCGTTTTACTTGTG-----GC------T-TAAAGTTAATTGGCGCTTTGTTGGCAGTCGCCGCCGCCTGCT--------------------------------------CACTATGTGGATTAAG |
| droWil2 |
scf2_1100000004963:3628956-3629251 - |
|
TTGA-ACGATTGGACATCGAGGGCTCAAGCACGACCAACGTGAAAATGAAT----GTAAGTA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AC-------------GGAAAACAAACAAAATTTTGT-----------GAAG--------------------------TG----ATAAAAAAAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CATATATATATATATAGA---TGAAACATCTGTAA----------ACTTTAGATATATATAAATATAT---------ATCAAAT-------------------------------------GATATTTGCGAAGGA---TTTTGTGACAATTT---G------------T-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTTTA---------------------------------------------ATAATTGTTG--TTGTT--TTT-TG----T---TTT---------------TTTTTTTGTTT----TATTGCATTTTCAAATGG-TGAAGTTGTTTGGAATCTCGTTGGCGTTTGC---CT--------------------------------------------TGTTATGTGGAATATG |
| dp5 |
XL_group1e:3878541-3878843 + |
dps_2675 |
TTGA-ACGGTTGGACATCGAGGGCTTAAGCACGACCAACGTGAAGATGAAT----GTAAGTG------------------CCAAA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGATAAGGATTG-CCAGTGAAATCAGTGACACGTTTAAGGA-------CCCAATATAGTGATCGATACAATACAATAC-----------------------AATA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CATACATAT--ATATACAAGA--------------------------------CACACATGTGTATATATTTT----------------------------------------------------------------TTTTCTGTGATTATTTGCTG------------T-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTTTC---------------------------------------------AAAACCGTTT--TCGTT--TTT-TT----TCCGTTTCGTTTTGGT--TT----------------------CGTTTCC----TG-TGAAGTTCTTTGGCGCCTTCTTTGCGCTCGC---CATCTGCG--------------------------------------CGACCTGTGGATTAAC |
| droPer2 |
scaffold_17:813733-814036 + |
|
TTGA-ACGGTTGGACATCGAGGGCTTAAGCACGACCAACGTGAAGATGAAT----GTAAGTG------------------CCAAA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGATAAGGATTG-CCAGTGAAATCAGTGACACGTTTAAGGA-------CCCAATATAGTGATCGATACAATACAATAC------------------------ATA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CATACATATATATATACAAGA--------------------------------CACACATGTGTATATATTTT----------------------------------------------------------------TTTTCTGTGATTATTTGCTG------------T-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTTTC---------------------------------------------AAAACCGTTT--TCGTT--TTT-TT----TCCGTTTCGTTTTGGT--TT----------------------CGTTTCC----TG-TGAAGTTCTTTGGCGCCTTCTTTGCGCTCGC---CATCTGCG--------------------------------------CGACCTGTGGATTAAC |
| droAna3 |
scaffold_13117:4704990-4705236 + |
|
GCTG-AGGCAACAACGA--------------------------------------------------------------------------------------------------CGG------TGGC-------------------GACTA-------------------------------------------------------------------------CTG---AGCTGA-TTTAATTAATAACGAAT-------------------------------------GCCAACTACAAGCAGAAGATCGATTTGAATGGC-------------AGAGAGCCCACAAGGAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------G--------------------------------------------------------------CAAGAACAAGAATAGAAAAAAAATAAAAA-----TAAAAAAATTAAA------------------------TCAAAA-ATAATAG----------AATTTGC-------------------------ATT-------------------------------------------------------------------------------------------------------------------AAAATACTTATGAATGATTACACAACAACA-------------------------------------------------------------------------------------------------------------------------------------------AAAAGTCAATTTGTTTTTGGT---------TGT-----------------------------------TCT--------------------------------------------------------------------TGTTGTTGTT---------------------------------------------------------GTTATTATTG |
| droBip1 |
scf7180000396006:47119-47400 - |
|
TTGT-ACGGTTGGAGATCGAA-GCTCAAGCACCACCGACTTAAAGATGAAT----GTAAGTG------------------------G------------TGCCCAC--ACAGGATTACC---CC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAAGGATTA-CTAAGGATATCTATAACTCA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------G-----------------------------------------T------------------------------------------------------GATA---------------------------------------------------------------------------------------------------------------------TAATT--------------------------------------------------------------------------------------ATCTTAATTTTTTTTCTGTGCAATAAAAAAAAACCAAAAAAGTTTCTAACAAAAAAAAAAACCAAAATTACAAA-CAA-CAAACA--------------------------------------------AAAA-------------------------------------------------------------------TGTTTTACTCGTT----------------------CGTTTCC----GGTTGAAGTTCTTTGGCGCCTTCATTGCGATCGC---CATCTGCG--------------------------------------GGATGTGTGGATTAAC |
| droKik1 |
scf7180000302486:2229277-2229509 - |
|
AGTG-CAAATGCA---------------------------------------------------------------------------------------GCAG-----------CAA------CAGC-------------------AGCAG-------------------------------------------------------------------------CAG---GCATCGTTTTCGTCGTTGTCGTCATCAAACCGAAAGAATGTATTCGGGCTGAGGAAAAT----TAATAAAAAAAAAAA----------------------------------------------------------------------------------------------------AAAGAGAGAAAAA------------------------------------------------------------------GAGAGGCGAA-------T-----------------------------------------------------------------------------------------------------------------------------------------------------------------GTGCTAA----------ATTTTAA------------------------------------------------------------------AAATATAT-----------A-----------------------------------------------------------------------------------------------------------------------------------------------------------------------CACTCGAAACCAGAAG------------------------AAGTGCTTACATTCATTTTTAT----TATTTC--------TTCTTGCC------------ATTT--TTGCACTT-----CTCAT---TTC-----------------------------------------------------------------------------------------TA--------------------------------------T---------TTCTAT |
| droFic1 |
scf7180000453843:644345-644550 - |
|
AAAA-TTCAAAGAACACCC-------------CAACAATCTAAAAAT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAACTAATTATTAGAATATATAATAAAAAAGTATACATAT------TAAAAAAGGAGG--------------------------------------------------------------------CATTAATTCGTATACATATT----------------------------------------------------------------TT--------------------------------------------------ACTGCTAACTTGC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTGTT--ATT-TA----TTACTCGT------AT--TTAT-------------GAAA---ACTTTTT----TG-TAAAGGTTACTAAATTTATCTT--------T---CATCTGCA--------------------------------------TCCT-TTTCAGTTACG |
| droEle1 |
scf7180000490564:1825011-1825224 - |
|
GAGC-CAC-A--------------------------------------------------------------------------------------------------GCAGCAGCAGC---AGAGAC-------------------ATTTG---CATAAGGCTC-TTTA---------------GAGATGGCAATGTGCGAAATATAAGTTGAGGCTGTAGGTGTTTCTAAACTTT-ACCAG---------------------AAA-----------------ATAAAAAATTTAA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTTTAA-------------------------------------------------------AGTAATACAAAAAAGAAACTAT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAATACTAACATTGGATTTAATTACCTGTTTCTTTTAAAT---------T-----------A--TATA--ATTA-----TTCAT---TTT---------------------------------------------------------------------------------------------------------------------------------------TTTGTTTGAA |
| droRho1 |
scf7180000780293:280510-280732 + |
|
CAAC-AAC-AACA---------------------------------------------------------------------------------------ACAG-----------CCA------TAAC-------------------AACAACAACAAACGAC---GACG---------------GCGAAGGCAA----------------------------------------------------------------------------------------------------------------CAAATAAAATCGAAAG-CCG-------------AAAGC-----------------------------------------------------------------------CGCG--TCCCAAAAAAACCGGTCCGT-CAATGTTCTA-----------------------------------------------------------------------------AAAATGAAACTGG-----------------------TTGCACCGAAAAAAAC--------AA-----------------------------------------------CAAG-------------------AACA-AAAACAAAAGGAG------------TACTAGACCAAAAA------------------------------------------------------TATATCTTTTT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TATTTT--------TTGTTGCA-------------A--TTTT--ATTT-----CTCCT---TTT--------------------------------------------------------------------------------------------------------------------------------------------TTTGT |
| droBia1 |
scf7180000302188:435583-435802 + |
|
CAAA-AAACAACAATC----------------------ACCAGGAGGCGAA----GACGC-A------------------GAAAAGCAAACGCAAGGCGCGCAAAT--ACAA------------A----------------------AGC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAAAT--AAAAACAAAA--------------------------------AACA-ATG--AGTGAAAAATTTCCCTG-----------------AAACACATTGACTTTTCAAAAAAATAA-------------------------------TACGACGAAGAGAAAATTCTCTAA----------------------------------------------TTTTTTTAA-------------------------------------------------------------------------------------------------------------------------------------------CATG--------TTTTTCCAACTTGT---T--AA--TTTT------------TCC----TCAC------AA--TTTT-------------GAAA--------------------------------TGA-------------------------------------------------------------------------TCTTT |
| droTak1 |
scf7180000415141:14313-14540 - |
|
CTAAACCGAGACC-----------------------ACAACAAAAGCCAAA----GCAAATG------------------CAAAAGC------------AAAAG--AAACAGCAGCAAGAACAGC----------------------AAAA----------------------------------GCTA-------------------------------------------------------------------------------------------------------------------------------ATGAAAG-AT----------------------------------AAACAACCAAGACCA---------------------------------------------------------------------------------AAACCAAA-----------------------------ACCAA-------AATCAAAACC----AA--G--TCCAAGAGTCAAGTTAG-----------------------AAGCACATGAAGAGCC--------------------------------------------------------------------------------------------------------------------------------------ATCAGAAATCAATTTTCAAATCAATCATTGCGCGATCCAAAAAATAA-------------------------------GAACAGAAGAACAAATTTCC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTTT |
| droEug1 |
scf7180000409182:66690-66895 + |
|
ACAA-TCGAGATG-----------------------AAAATGAAAACGAAT---------TG------------------------C------------AGGAGCA--GCAGAAGCAGCAGCAGCAGC-------------------AG----------------------------------------------------------------------------CAG-----------------------------------------------------------------------------------------------GC-------------AAAAAGCTTACGAAACTAATT----------------------------------------------------------------------GCAGCTGGTTTATGCACTGAACAAAAAGCAAAGGTCATTAAAACCAAATAAAAAATCAAGAAAATCAAA-AGTAAAAATTAAAAAA----AA--G--T----------------------------------------------------------------------------------------------------CCTTTTAAA------------------------ATA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AA---CT--------------------------------------------TCTTATATAATCTAAT |
| dm3 |
chrX:14617507-14617792 - |
|
AAAT-GAA------------------------------------GAAAAAT----GCGAGGG-------------------------------------------------------A------CGCC-------------------AAC-------------------------------------------------------------GGGAGACGCCGGTGCTC---AACTAA-ATTGA---------------------AAA-----------------ACAAAAAGGTTAACTATAAAAAAAAAATCA----------------------------------------------------------------------------------------------------AAGCAAAA------------------------------------------------------------------AGAAAATAAA-------AATCCCAAAC----AA--A--A--------------------------------------------------------------CGAGAGGAAGAACAAAGCAGCAGCAACGA-----CGGGGCAGCAACG------------------------TCAACC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAATTTCACGCTGACAAATTATGATTTATGCTACAGCTG-------------------------------------------ACTGCTG--CCGTT--TGG-TT------CTTCTTGTTTGAGT--TT----------------------CAGTTTT----TG-TTT-GGATGCGGGCTTTTTTTTTGCTTTTGT------------------------------------------------------------TTCTG |
| droSim2 |
3l:12126171-12126412 - |
|
TTCA-GCGAGTCGGCGGG---CGTTG----------AGTTTGAGTGCGAAGTGCGGTGCGCGTCGAGTCAGCGAGCGTTGGTTGAGC------------AGCA-----GCAGCAGTAGC---AGCAGC-------------------AACAA---CACGACGCAACAGCAACATCAGCCGCAACAGCAG-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGCAACAGA-AGCA------GCAACTTGCAACTTGCAACAGCAACACAGGCGAG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGGTT-------------------------------------------CCGGTGCGCGTTTTCGAGTGGCGTGCCAAAAAACAAAAAAAAAAGTTCACAA------ATAA |
| droSec2 |
scaffold_6:3200758-3200930 + |
|
CCGG-CCGACACAAAC------ACACTCACACAGCCAAGCGGTAACTTCGA----AAAAAAG------------------------C------------AGCGG--CCGCAGCAGCAGCAGCGCC----------------------AACAA---CTAAGCGA-------------------------------------------------------------------------------------------------------------------GCAAAAAAAAAA----AAATCAAATACAACA----------------------------------------------------------------------------------------------------AAAAAAGGACA--T-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AG--CAAA----------------------------------------------------------------AAAACCGAAAAAATATC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------CAAACGCA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CACAGAGAAAAAAG |
| droYak3 |
X:14958994-14959283 + |
|
TTGA-TCGGTTGGACATCGAGGGCTCAAGCACGACCACTGTGAAGATGAAT----GTAAGCGCTGCACCTGCCCCC------CAT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGCTAAGGATTA-CC----------------------------------ATTCAGCCAA-------------------GGATTCATTCACCACAC------------------A-----------------------------------------------CA------AAA------------------------------------------------------------TTCCACATCCATCCACATAACAG-----------------------------------------T------------------------------------------------------GATA-AT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATAACAACATATCTATAACATCTT--TTTTTTTTCTGT-----------------------------------------------------------------------------------------------GACTC---------------------------------------------AAA---GTAT--TTGTT--TTG-TG----TTTGTTTCCTTAC-GA--TT----------------------CGTTCTC----GG-TGAAGTTCTTTGGCGCCTTCATTGCGCTCGC---CATCTGTG--------------------------------------CGCTATGCGGATTAAC |
| droEre2 |
scaffold_4845:21006488-21006703 - |
|
AGTT-GCGAAAACA--------------------------------------------------------------------------------------GAAA-----------C------ATTAGATTTTTCATTTAATAACTGCAAC---GGCT-AAGGC---GATG---------------GCGACGGCGA------------------------------CT----------------------------------CGAAAA--------------------------------------------AATTGTCAACTT-CC----------------------------------TGTTGGACAA-------------------AAGCGAAAAAACGAAAA------------------A-----------------------------------------------CA------AAA------------------------------------------------------------AACAAAAAAAGGAAAAATAATGA-----------------------------------------AGAAAAGAAGA-----AAATGCAACTGAG------------------------TAAGAC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AACC----------------------------------------CACTAATTGCCTCATTTCGGT---------C-----------G--TGTG--T----------------T---------------------------------------------------------------------------------------------------------------------------------------------TTCTTT |