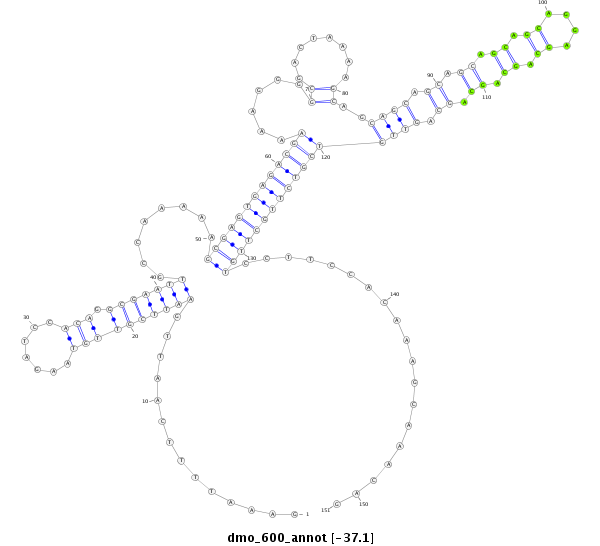
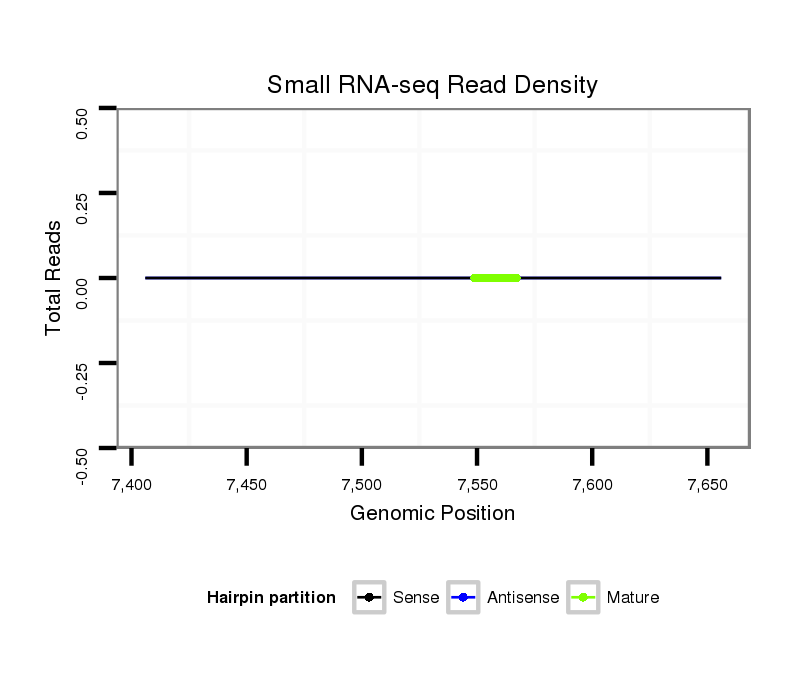
| GTGAAGTTG-----AAAC-TGGCACAATTGGATCGCCGTCGTCGTCAGCA---------------------------------------------------------------AAT-------------------------------CAAAATCCAAA-CCCAAAT-------CATTTAAAAACGAAATGCAAACC-------G------GG---ACAG-AGCGTTG-----------------------------------------------------------------------------------------------------------------------------------CTGAGGGCAAGCCTTCAGCGAAAGAGACCA--------------------------------------------------------------TCGTCG---ATAAC------------------------AGCAAGAGCTGCAGCAAAACCAAGA--------------------------------------------------------ATATT--------------------------------------------------------------TC---------------------------C---------------------------------------------GATAAAT-TGCAGAGCAACA-------AGTTT-----ATAATTC---AACAGCAGCAGCAGCAACAAC------------------------------------------------AACAA | Size | Hit Count | Total Norm | Total | M026
Head | M043
Female-body | M056
Embryo | V046
Embryo | V052
Head | V058
Head | V120
Male-body | SRR1275488
Male larvae | SRR1275486
Male prepupae | SRR1275484
Male prepupae | GSM1528802
follicle cells |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ATT--------------------------------------------------------------TC---------------------------C---------------------------------------------GATAAAT-TGCAGAGCAACA-------AGT........................................................................................... | 28 | 1 | 3.00 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................GCAAACC-------G------GG---ACAG-AGCG................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 18 | 1 | 3.00 | 3 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................GCAAACC-------G------GG---ACAG-AGCGTTG-----------------------------------------------------------------------------------------------------------------------------------CTGA...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 2.00 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................AACC-------G------GG---ACAG-AGCGTTG-----------------------------------------------------------------------------------------------------------------------------------CTGA...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 2.00 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................G---ACAG-AGCGTTG-----------------------------------------------------------------------------------------------------------------------------------CTGAGGGC.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 1 | 2.00 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................ATGCAAACC-------G------GG---ACAG-AGC................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................CAAACC-------G------GG---ACAG-AGCGTTG-----------------------------------------------------------------------------------------------------------------------------------CTGAGGGCA................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................AACGAAATGCAAACC-------G------GG---ACA...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................GGGCAAGCCTTCAGCGAAAGA................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ATATT--------------------------------------------------------------TC---------------------------C---------------------------------------------GATAAAT-TGCAGAGC......................................................................................................... | 23 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................AACC-------G------GG---ACAG-AGCGTTG-----------------------------------------------------------------------------------------------------------------------------------CTGAG..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................ACC-------G------GG---ACAG-AGCGTTG-----------------------------------------------------------------------------------------------------------------------------------CTGA...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................ACC-------G------GG---ACAG-AGCGTTG-----------------------------------------------------------------------------------------------------------------------------------CTGAGGGC.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................GCAAACC-------G------GG---ACAG-AGCGTTG-----------------------------------------------------------------------------------------------------------------------------------CTGAGG.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AACA-------AGTTT-----ATAATTC---AACAGCAGC................................................................. | 25 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AT-TGCAGAGCAACA-------AGTTT-----ATAATT.............................................................................. | 25 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................CC-------G------GG---ACAG-AGCGTTG-----------------------------------------------------------------------------------------------------------------------------------CT........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................GGGCAAGCCTTCAGCGAAAGAGACCA--------------------------------------------------------------TC............................................................................................................................................................................................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................T-TGCAGAGCAACA-------AGTTT-----ATAATT.............................................................................. | 24 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................GCAAACC-------G------GG---ACAG-AGCGT............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................G---ACAG-AGCGTTG-----------------------------------------------------------------------------------------------------------------------------------CTGAGG.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................AGCCTTCAGCGAAAGAGACCA--------------------------------------------------------------TCGTC......................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAAGAGCTGCAGCAAAACCAAG................................................................................................................................................................................................................................................................................................................................ | 22 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AAT-TGCAGAGCAACA-------AGTTT-----ATAATT.............................................................................. | 26 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................G------GG---ACAG-AGCGTTG-----------------------------------------------------------------------------------------------------------------------------------CTGA...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................................................................CG---ATAAC------------------------AGCAAGAGCTGCAGC......................................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................AATGCAAACC-------G------GG---ACAG-AGCGTT.............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................ATGCAAACC-------G------GG---ACAG-AG.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 18 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................AACGAAATGCAAACC-------G------GG---ACAG-AGCG................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................CC-------G------GG---ACAG-AGCGTTG-----------------------------------------------------------------------------------------------------------------------------------CTGAGGGC.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................ATTTAAAAACGAAATGCAAACC-------G------GG---AC....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................................GACCA--------------------------------------------------------------TCGTCG---ATAAC------------------------AGCAAGA................................................................................................................................................................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................ACAG-AGCGTTG-----------------------------------------------------------------------------------------------------------------------------------CTGAGGG................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................GAAAGAGACCA--------------------------------------------------------------TCGTCG---ATAA................................................................................................................................................................................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................GAAATGCAAACC-------G------GG---ACAG-AGC................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................................GAGACCA--------------------------------------------------------------TCGTCG---ATAAC------------------------AGC..................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ATAAAT-TGCAGAGCAACA-------AGT........................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AACA-------AGTTT-----ATAATTC---AACAGCAGCAGC.............................................................. | 28 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................AGCGTTG-----------------------------------------------------------------------------------------------------------------------------------CTGAGGGCAAGC.............................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................GCAAACC-------G------GG---ACAG-AGCGTTG-----------------------------------------------------------------------------------------------------------------------------------CTG....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................CAAGCCTTCAGCGAAAGAGACCA--------------------------------------------------------------TC............................................................................................................................................................................................................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................CC-------G------GG---ACAG-AGCGTTG-----------------------------------------------------------------------------------------------------------------------------------CTGAGA.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GCAAGAGCTGCAGCAAAACCAAGA--------------------------------------------------------....................................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGCAGCAGCAGCAACAAC------------------------------------------------AAC.. | 21 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |